CYP450 Metabolites Analysis Service
Creative Proteomics specializes in high-precision profiling of CYP450-mediated metabolism. We help you quantify oxidative metabolites, uncover pathway mechanisms, and compare metabolic behavior across species and matrices.
Whether you're working with drug candidates, environmental chemicals, or endogenous substrates—we deliver the analytical clarity you need.
What Sets Our Service Apart?
- Targeted + Exploratory Coverage
Absolute quantification of key metabolites plus MS/MS support for unknown identification - Isoform-Specific Resolution
Selective profiling for CYP1A2, 2D6, 3A4 and more using recombinant or microsomal systems - Multi-Matrix Compatibility
Validated for plasma, urine, liver fractions, cell cultures, bile, and tissue homogenates - High Analytical Confidence
CV<5%, LOD down to 1 nM, and full integration with pathway mapping tools
Submit Your Request Now
×
What You Will Receive
- Quantified CYP450 Metabolite Panel
Absolute concentrations for all targeted analytes (e.g., 6β-OH-testosterone, Nortriptyline) - Raw + Normalized Data Tables
With QC metrics, retention time, internal standard recovery - Pathway Mapping Summary
Visual metabolite conversion flow (e.g., CYP3A4-mediated hydroxylation → glucuronidation) - Custom Interpretation Report
Biological relevance + compound-specific insights from experienced analysts
- What We Provide
- Advantages
- Technology Platform
- Sample Requirements
- Demo
- FAQs
Overview of Cytochrome P450 and Its Role in Metabolism
Cytochrome P450 (CYP450) enzymes form a superfamily of heme-containing monooxygenases that catalyze the oxidative metabolism of a wide range of endogenous compounds and xenobiotics. These enzymes are central to phase I metabolic processes, introducing functional groups that increase solubility or prepare substrates for downstream conjugation. Key reactions include hydroxylation, dealkylation, epoxidation, and sulfoxidation. Each CYP450 isoform—such as CYP1A2, CYP2D6, and CYP3A4—displays distinct substrate preferences and expression profiles across species and tissues. Understanding CYP450 activity is essential for deciphering how compounds are modified and eliminated by biological systems.
Why Analyze CYP450 Metabolites?

Reveal Pathways
Identify oxidative reactions such as hydroxylation, dealkylation, or epoxidation to map primary metabolic routes.

Assess Stability
Quantify how fast a compound is metabolized and estimate its biological half-life.

Compare Species
Analyze CYP isoform activity across species (e.g., human, mouse, dog) to support translational research.

Monitor Accumulation
Detect minor or slowly cleared metabolites that may accumulate in tissues or fluids.

Study SMR
Correlate chemical structure with metabolic behavior to guide compound design.

Map Pathways
Reconstruct CYP450-involved pathways using quantification and structural data.
CYP450 Metabolites Analysis Service Offered by Creative Proteomics
- Quantitative Analysis of CYP450 Metabolites: Absolute quantification of oxidative metabolites using LC-MS/MS and/or GC-MS platforms, with internal standard calibration for high accuracy.
- Isoform-Specific Metabolism Profiling: Analysis of metabolic products generated by individual CYP isoforms (e.g., CYP1A2, CYP2D6, CYP2C9, CYP3A4) using recombinant enzymes or isoform-enriched microsomes.
- Time-Course Metabolite Kinetics: Monitor metabolite formation over time to assess reaction rates, turnover kinetics, and stability of the parent compound.
- Species Comparison Studies: Comparative metabolism studies using liver microsomes or hepatocytes from human, rat, mouse, dog, or monkey sources.
- Metabolic Hotspot Identification: Structural elucidation of oxidation-sensitive sites using tandem MS fragmentation and metabolite mapping.
- Metabolite Pathway Reconstruction: Integration of detected metabolites into KEGG/BioCyc pathway maps, enabling visualization of primary and secondary metabolic routes.
- Reactive Metabolite Detection (Optional): Use of trapping agents such as glutathione (GSH) to detect electrophilic or unstable intermediates formed via CYP450 catalysis.
- Matrix Versatility: Analysis of metabolites in plasma, serum, urine, bile, liver microsomes, S9 fractions, cell culture supernatants, and tissue homogenates.
List of Detected CYP450 Metabolites and Related Compounds
| Metabolite Class | Example Compounds | Metabolic Pathways |
|---|---|---|
| Hydroxylated Metabolites | 4-Hydroxybiphenyl, 6β-Hydroxytestosterone | CYP1A2, CYP2C9, CYP3A4-mediated oxidation |
| N- and O-Dealkylated | Nortriptyline, Desmethylcitalopram | CYP2D6, CYP3A4 |
| Epoxide Intermediates | Aflatoxin B1-8,9-epoxide, Benzo[a]pyrene epoxide | CYP1A1, CYP3A4 |
| Sulfoxide/Amine Oxides | Sulindac sulfone, Clozapine N-oxide | CYP3A4, CYP2D6 |
| Desaturation Products | Tretinoin, Retinoic acid isomers | CYP26A1, CYP3A7 |
| Glucuronidated/Sulfated | 4-hydroxybiphenyl sulfate | Combined Phase I/II metabolism |
| Endogenous Substrates | Testosterone, Estradiol, Cholesterol derivatives | CYP19A1, CYP11A1, CYP27A1 |
Custom panels available based on compound structure or species.
Advantages of CYP450 Metabolites Assay
- Quantification CV<5%: High reproducibility across replicates with isotope-labeled internal standards.
- LOD as low as 1 nM: Ultra-sensitive LC-MS/MS detection suitable for low-biomass or CSF samples.
- CYP450 Metabolites/ATP/ADP ratio with<10% error: Reliable energy profiling in serum, liver, and cell lysates.
- Compatible with 10+ sample types: Validated for plasma, tissues, urine, CSF, and cell-based matrices.
- 20+ related metabolites covered: Broad profiling of purine metabolism and CYP450 MetabolitesK-linked pathways.
Methods and Instrumentation for CYP450 Metabolites Analysis
Agilent 6495C Triple Quadrupole LC-MS/MS – Ultra-sensitive MRM-based quantification of known metabolites
Agilent 7890B GC system with 5977A MSD – High-resolution GC-MS analysis of volatile and derivatized metabolites
Agilent 1260 Infinity II HPLC – High-stability front-end separation for complex metabolic mixtures

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Agilent 7890B-5977A (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Workflow for CYP450 Metabolites Analysis Service
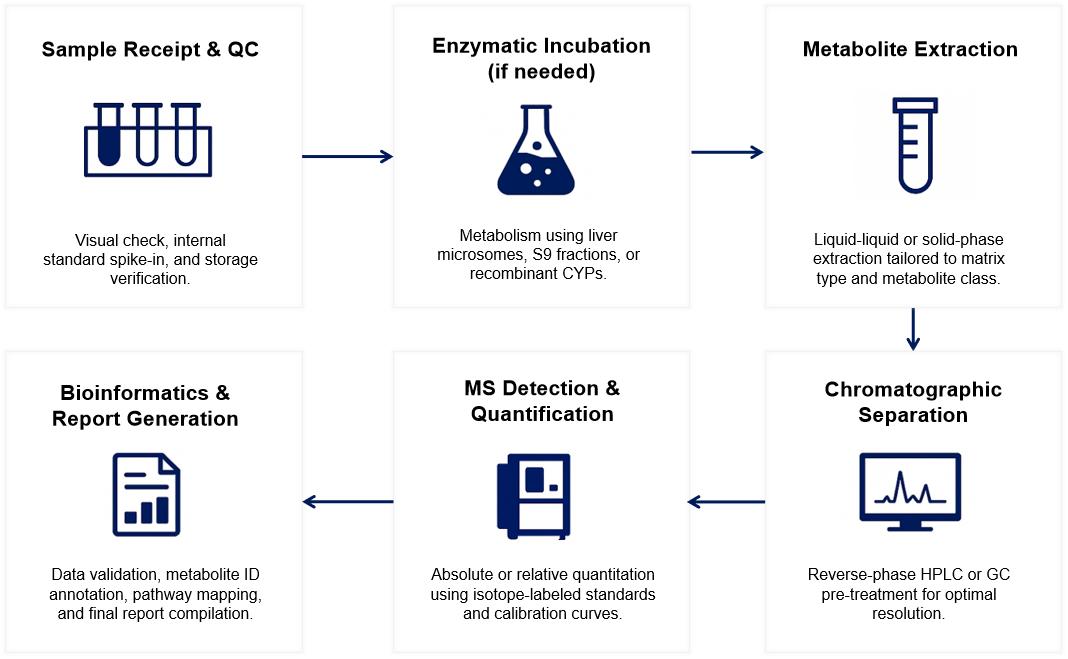
Sample Requirements for CYP450 Metabolites Analysis Service
| Sample Type | Minimum Amount Required | Storage & Transport Conditions | Remarks |
|---|---|---|---|
| Plasma / Serum | ≥ 200 µL | -80°C, avoid freeze-thaw cycles | Anticoagulant info (e.g., EDTA) recommended |
| Liver Microsomes | ≥ 0.5 mg protein | -80°C, aliquoted if possible | Specify species and preparation method |
| Hepatocyte Suspension | ≥ 1 × 10⁶ cells | Cryopreserved in liquid nitrogen or -150°C | Include viability data if available |
| S9 Fractions | ≥ 0.5 mg protein | -80°C, protease inhibitor optional | Useful for phase I + II studies |
| Cell Culture Medium | ≥ 500 µL | -20°C, tightly sealed | Indicate compound concentration + exposure time |
| Tissue Homogenates | ≥ 100 mg | Snap-frozen, no preservatives | Homogenization buffer info optional |
| Urine / Bile | ≥ 500 µL | -80°C, aliquoted to prevent degradation | For excretion/metabolic clearance analysis |
| Pure Compounds / NCEs | ≥ 10 µg (solid or in solvent) | RT or -20°C depending on stability | Indicate solvent (DMSO, ACN, etc.) and purity |
Demo Results
FAQ of CYP450 Metabolites Analysis Service
Can I submit samples that have already been incubated with CYP enzymes?
Yes. We accept pre-incubated samples as long as sufficient documentation is provided, including incubation time, enzyme source, cofactors used, and compound concentration.
Is it possible to analyze metabolites in both parent compound-spiked samples and untreated controls?
Absolutely. We recommend including matrix-only controls and parent-only samples to help distinguish endogenous signals, baseline drift, or matrix interferences during data interpretation.
Can Creative Proteomics help identify unexpected or novel metabolites?
Yes. In addition to targeted panels, we offer extended MS/MS screening for unknown metabolite discovery, supported by structure-based prediction and diagnostic fragmentation interpretation.
How should I prepare liver microsome or S9 fraction samples for submission?
We recommend submitting snap-frozen aliquots (no additives), stored at –80°C, with clear labels indicating protein concentration, species origin, and preparation date.
Do you support metabolite analysis in co-culture or 3D cell systems?
Yes. We can analyze metabolites generated in conditioned media from co-cultures, organoids, or 3D systems, provided sufficient volume and matrix information are available.
How are background signals or matrix effects handled during analysis?
Matrix-specific blanks and spike-in recovery checks are used to identify and correct for potential suppression or enhancement effects during quantification.
Can metabolite formation be monitored over time (kinetics)?
Yes. We support multi-timepoint analysis (e.g., 0, 5, 15, 30, 60 min) for kinetic profiling, allowing rate constant estimation and metabolic half-life calculation.
What if my test compound is light-sensitive or unstable?
We recommend protecting samples from light during handling and clearly noting stability constraints on the sample submission form. We also offer cold-chain processing during sample prep.
Do you provide visual summaries for complex metabolite profiles?
Yes. Upon request, we can deliver heatmaps, chromatographic overlays, and pathway diagrams to support data interpretation and presentation.
Can I request isotope-labeled internal standards for specific metabolites?
Yes. Where available, we include isotope-labeled standards for absolute quantification. For custom compounds, we can assist with sourcing or recommend suitable surrogates.
Learn about other Q&A.
Publications
Here are some of the metabolomics-related papers published by our clients:

- Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy. 2023. https://doi.org/10.1111/nan.12931
- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023. https://doi.org/10.1038/s42003-023-04739-9
- The activity of the aryl hydrocarbon receptor in T cells tunes the gut microenvironment to sustain autoimmunity and neuroinflammation. 2023. https://doi.org/10.1371/journal.pbio.3002000
- Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. 2023. https://doi.org/10.1038/s41556-023-01162-4
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023. https://doi.org/10.1101/2023.11.09.566426
- Anti-inflammatory activity of black soldier fly oil associated with modulation of TLR signaling: A metabolomic approach. 2023. https://doi.org/10.3390/ijms241310634
- Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. 2023. https://doi.org/10.3390/plants12030651









