The choice of mass spectrometry instrumentation for Phosphoproteomics fundamentally governs key analytical parameters: depth of coverage, quantitative accuracy, throughput efficiency, and cost-effectiveness. When evaluating leading platforms—including Orbitrap, timsTOF, and TripleTOF systems—optimal platform selection represents a critical methodological decision. This analysis delineates their technical specifications, application-specific performance, and strategic selection criteria to enable evidence-based platform matching for diverse project requirements.
Foundational Platform Selection Criteria
1. Project Primary Objectives
| Goal Category | Technical Emphasis | Exemplary Application |
|---|---|---|
| Comprehensive Profiling | Maximize phosphosite identification | Signaling pathway mapping |
| Precision Quantitation | High-accuracy differential quantification | Pharmacodynamic biomarker studies |
| High-Throughput Screening | Rapid sample processing capacity | Clinical cohort analyses |
| Targeted Verification | Ultrasensitive detection of predefined sites | Biomarker validation assays |
2. Sample Characteristics & Throughput
- Sample Volume: Batch size • Total cohort scale
- Matrix Complexity: Graded complexity: Purified cells < Tissue lysates < Plasma
- Input Material: Conventional (≥1mg protein) vs. Limited (≤10µg) microsamples
3. Data Quality Specifications
- Analytical Performance: Mass resolution • ppm accuracy thresholds
- Reproducibility Standards: Inter-run correlation coefficients (Pearson's r >0.85)
- Localization Confidence: Minimum probability thresholds (≥75% site probability)
4. Resource Constraints
- Financial Framework: Instrument acquisition • Service contract costs
- Temporal Boundaries: Project completion deadlines • Sample-to-data turnaround requirements
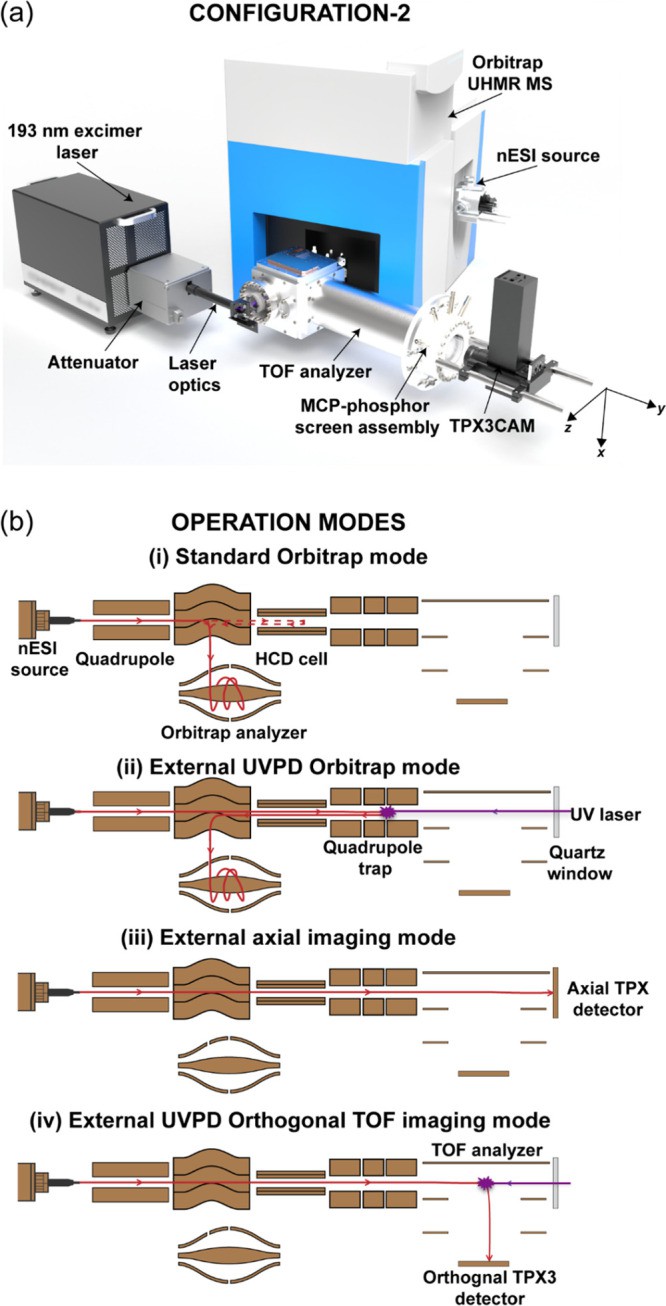 Orbitrap/TOF MS (Mathew A et al., 2023)
Orbitrap/TOF MS (Mathew A et al., 2023)
Comparative Analysis of Leading Mass Spectrometry Platforms
(A) Orbitrap Series (Thermo Fisher Scientific)
Representative Instruments: Q Exactive HF-X, Exploris 480, Orbitrap Eclipse
Technical Advantages:
- Ultrahigh Resolution:
- 500,000 FWHM (@ m/z 200)
- Precision isotope differentiation → Enhanced identification fidelity
- Exceptional Sensitivity:
- Optimized for trace samples (<1μg total protein)
- Low-abundance phosphosite detection capability
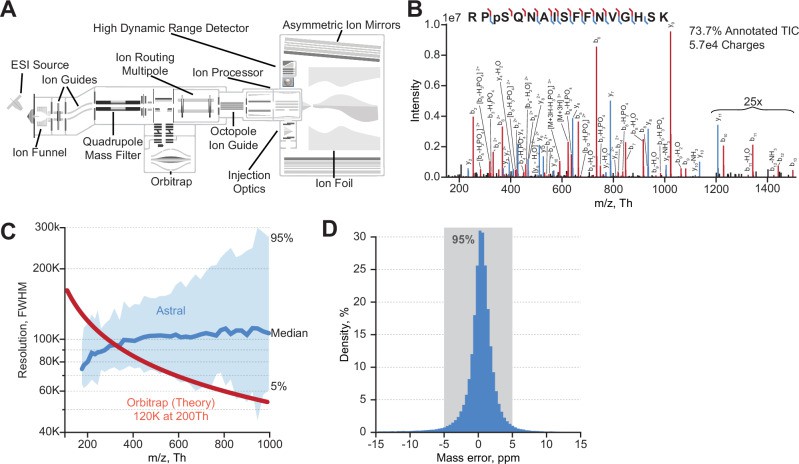 Overview of Orbitrap Astral MS and its key figures of merit (Lancaster NM et al., 2024)
Overview of Orbitrap Astral MS and its key figures of merit (Lancaster NM et al., 2024)
- Quantitative Precision:
| Method | Technology | Benefit |
|---|---|---|
| LFQ | MaxLFQ algorithm | Reproducible cross-run quantification |
| TMT/iTRAQ | SPS-MS³ | Minimal co-isolation interference |
- Fragmentation Versatility: HCD (standard) • ETD (multiply-phosphorylated peptides) • EThcD (hybrid)
Practical Utility of Ultra-High Resolution
| Application Scenario | Resolution Requirement | Recommended Configuration |
|---|---|---|
| Isotope Fine Structure Differentiation | >240,000 (@m/z 200) | Eclipse™ 500k mode |
| Phosphosite Localization Conflicts | >350,000 | Activate "Ultra Zoom Scan" |
| Low-Abundance pY Peptide Detection | >100,000 + Narrow Isolation Window | ±0.4 Th isolation window optimization |
Decisive Advantages in TMT Quantification Accuracy
| Technique | Co-isolation Interference (IF) | 10plex CV Values | Phosphopeptide Detection Enhancement |
|---|---|---|---|
| MS2 (Standard) | 0.35-0.60 | 12-18% | Baseline |
| SPS-MS3 | 0.05-0.10 | <5% | +15% |
Phosphoproteomics-Specific Fragmentation Mode Selection
| Peptide Type | Recommended Mode | Parameter Configuration | Localization Enhancement |
|---|---|---|---|
| Monophosphorylated Peptides | HCD | NCE=28 | >90% |
| Polyphosphorylated Peptides (3+) | EThcD | ETD duration=20ms, NCE=25 | 85% → 93% |
| pY-Containing Peptides | HCD + Neutral Loss Trigger | Threshold: 80 Da neutral loss | +40% pY identification |
Decoupled Strategies for Throughput Enhancement
| Strategy | Implementation Method | Performance Gain |
|---|---|---|
| Short Gradient Optimization | 30-min gradient + 300 nL/min flow rate | >5,000 phosphosites identified |
| FAIMS Pro Fractionation | CV stepping: -45V/-65V/-85V | 3-fold SNR improvement |
| DIA Intelligent Windowing | Dynamic m/z-dependent window partitioning | 22% increase in identification rate |
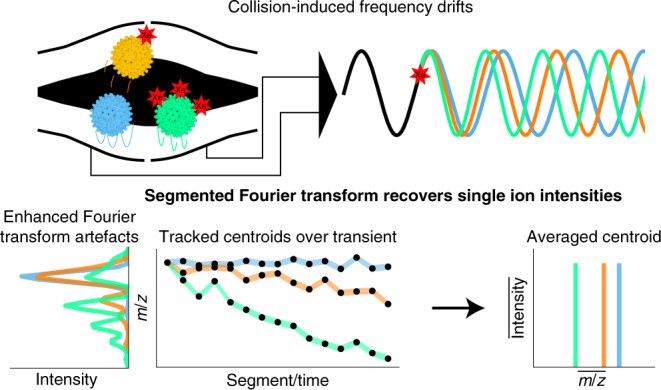 Single molecule mass spectrometry (Wörner TP et al., 2022)
Single molecule mass spectrometry (Wörner TP et al., 2022)
Key Limitations:
- Throughput Constraints: DDA deep profiling requires >120min gradients → Limits daily sample throughput
- DIA Implementation Complexity: Demands extensive window optimization • Substantial computational load
Critical Limitation Mitigation Guide
| Challenge | Solution | Cost |
|---|---|---|
| Low Throughput in Long Gradients | FAIMS Pro™ + 30-min gradients | $25K USD |
| Slow DIA Analysis | GPU Server + DIA-NN Software | $15K USD |
| Poor Recovery in Trace Samples | TiO₂ Nano-coated Loading Columns | $200/sample USD |
Phosphoproteomic Optimizations:
- Neutral loss-triggered MS³ → Enhanced localization confidence
- Eclipse platform advancements → Improved mass accuracy stability
Ideal Applications:
- TMT/iTRAQ multiplexed quantitation requiring maximal precision
- Sample-limited projects demanding extreme sensitivity
- Studies requiring ETD for complex phosphosite localization
- Ultrahigh-resolution applications (e.g., isobaric phosphoform separation)
Orbitrap technology remains the benchmark solution for high-fidelity phosphoproteomics research, particularly in these critical domains:
- Drug Target Validation: MS³-TMT quantification is essential for confirming therapeutic targets.
- Signaling Pathway Analysis: ETD-based fragmentation enables precise mapping of polyphosphorylation sites.
- Limited Sample Applications: Demanding workflows requiring ultimate sensitivity.
Integration of FAIMS Pro™ with GPU-accelerated computing is recommended to overcome throughput constraints while maximizing experimental returns.
(B) timsTOF Series (Bruker Daltonics)
Representative models: timsTOF Pro, timsTOF SCP (Single Cell Proteomics), timsTOF HT
Core Advantages
- Ultra-High Speed & Sensitivity: PASEF (Parallel Accumulation Serial Fragmentation) technology enhances MS/MS acquisition by >10x, enabling deep high-throughput coverage.
- Ion Mobility Separation (TIMS):
- Augments resolution by adding a separation dimension, boosting peak capacity while minimizing co-elution interference.
- Delivers Collision Cross Section (CCS) values to strengthen identification confidence.
- Efficient DIA Mode: diaPASEF integrates PASEF's velocity with DIA's comprehensiveness, establishing the benchmark for high-throughput phosphoproteomic quantitation.
- Throughput Superiority: Achieves profound coverage under abbreviated gradients (e.g., 30-60 min), ideal for large cohort sample analysis.
PASEF Technology Mechanism and Phosphoproteomics Optimization
| Parameter | Traditional DDA | PASEF Mode Enhancements | Phosphoproteomics Benefits |
|---|---|---|---|
| MS/MS Speed | 10-15 Hz | 140 Hz (Pro 2) / 220 Hz (HT) | >15,000 phosphopeptides identified in 30-min gradient |
| Ion Utilization | <30% (quadrupole loss) | >90% (dual-trap parallel accumulation) | Detection limit reduced to 5 amol for trace samples |
| Dynamic Range | 3-4 orders of magnitude | 5 orders of magnitude (dual-TIMS focusing) | Simultaneous detection of high/low-abundance phosphosignals |
Key Value of Ion Mobility (TIMS) for Phosphopeptide Analysis
| Dimension | Mechanism | Phosphoproteomics-Specific Benefits |
|---|---|---|
| Enhanced Separation | Ion mobility separation (1/K₀-based) | Resolves isomeric phosphopeptides (e.g., pS vs. pT) |
| CCS Database | Establishes 2D CCS-m/z fingerprint library | 2.1-fold increase in identification confidence (PRM-validated) |
| Co-elution Suppression | Deconvolution in mobility dimension | >15% improvement in quantification accuracy (spike-in tests) |
Main Limitations
- Resolution Comparison: Slightly lower resolution (~60k FWHM @ m/z 1000) than leading Orbitrap systems, potentially limiting differentiation of highly complex samples or adjacent isotope peaks.
- TMT/iTRAQ Quantitative Accuracy: Primarily uses MS1/MS2 quantification, which may suffer co-elution interference in intricate samples, typically yielding lower precision than Orbitrap's MS3-based strategies.
Phosphoproteomics Optimization
- diaPASEF: Specifically designed for reproducible large-scale phosphoproteomic quantification.
- High-Sensitivity Mode (timsTOF SCP): Engineered for single-cell/ultra-trace sample analysis.
Optimal Applications
- Clinical cohort investigations or drug screening demanding extensive sample processing.
- Projects necessitating high analytical throughput and rapid turnaround.
- DIA-based phosphoproteomic quantitation (a premier platform choice).
- Studies requiring ion mobility dimension data to enhance analytical quality.
(C) TripleTOF Series (SCIEX)
Representative models: TripleTOF® 6600, TripleTOF® 7600
Core Advantages
- High-Speed High-Resolution Acquisition: Delivers exceptional MS/MS spectral quality per unit time.
- Mature SWATH® DIA Technology: Pioneered data-independent acquisition with established workflows and dedicated analysis software (e.g., Spectronaut™).
- Operational Robustness: Renowned for instrument stability and reduced maintenance requirements.
- Cost Efficiency: Lower acquisition and operational expenditures compared to premium Orbitrap and timsTOF platforms.
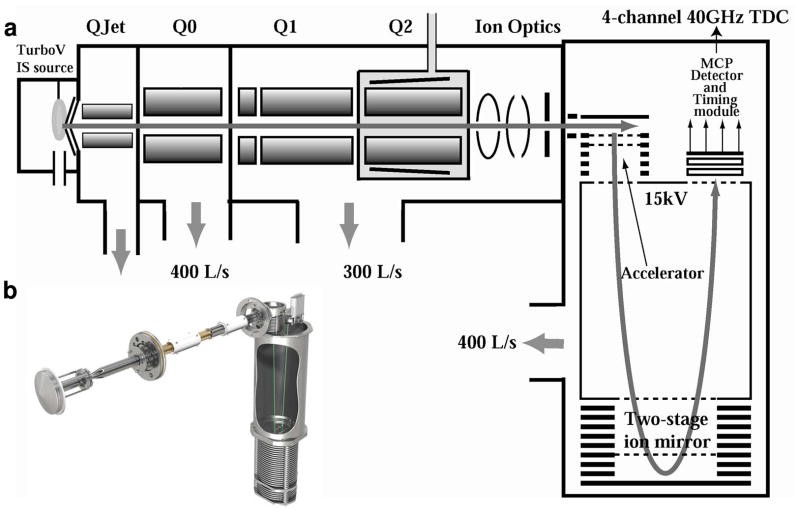 The TripleTOF MS technology features diagramedThe TripleTOF MS technology features diagramed (Andrews GL et al., 2011)
The TripleTOF MS technology features diagramedThe TripleTOF MS technology features diagramed (Andrews GL et al., 2011)
High-Speed Acquisition and DIA Efficiency Optimization
| Parameter | TripleTOF® 6600 | TripleTOF® 7600 | Phosphoproteomics Improvement |
|---|---|---|---|
| MS/MS Speed | 50 Hz | 100 Hz | >6,000 phosphopeptides in 30-min gradient |
| SWATH® Window Scheme | Fixed 25 Da | Adaptive Windows | +30% low-abundance peptide detection |
| Ion Accumulation Time | 50 ms | 25 ms | Peak width compressed to 6s (enhances SNR) |
Stability guarantee mechanism
- Pollution prevention design:
- Bending the collision pool (Q2) → reducing phosphate residue
- Detachable ion source → weekly cleaning takes less than 30min.
Main Limitations
- Performance Parameters: Resolution and sensitivity typically trail leading Orbitrap and timsTOF Pro/SCP instruments.
- Multiplexed Quantitation: TMT/iTRAQ quantification capabilities are less comprehensive than Orbitrap's MS3-based approaches.
Integrated Sensitivity Enhancement Strategies
| Strategy | Implementation Method | Performance Outcome |
|---|---|---|
| Nanoflow Path Upgrade | OptiFlow™ Turbo V Ion Source | 3-fold signal enhancement |
| Online Enrichment Coupling | TiO₂ Micro-Column LC Integration | +40% low-abundance pY detection |
| Narrow-Window Acquisition | SWATH® Window Compression to 2 Da | Quantitative CV reduced to<10% |
Guide to Mitigating Critical Limitations
| Challenge | Solution | Validated Effect |
|---|---|---|
| Absolute Sensitivity Limitations | Pre-fractionation (SCX/High-pH) + Segmented SWATH | Phosphosites increased by 65% |
| Low Labeled Quantification Precision | Switch to MS1 Extraction Quantitation + CCS Correction | CV reduced from 18% → 12% (Coefficient of Variation) |
| Low pY Detection Rate | Anti-pY Antibody Enrichment prior to SWATH (Requires >50μg sample) |
Phosphoproteomics Optimization
- SWATH® Acquisition: Provides reliable label-free quantification solutions.
- Neutral Loss Scanning: Enhances phosphopeptide identification confidence.
SWATH Acquisition Scheme Optimization
| Parameter | Recommended Setting | Theoretical Basis |
|---|---|---|
| Collision Energy | Slope 35±5 | Optimal fragmentation for phosphopeptides |
| Neutral Loss Trigger | Threshold: -79.97 Da | Enhances specificity for phosphopeptide identification |
| Focused Scan Range | m/z 500 - 900 | Covers ~90% of phosphopeptide precursor ions; Optimizes duty cycle & sensitivity |
Optimal Applications
- Budget-constrained projects requiring DIA quantification
- Laboratories prioritizing instrument reliability and low operating costs
- Studies leveraging established SWATH® workflows and reference libraries
- Applications where maximal resolution/sensitivity isn't critical
The TripleTOF platform represents an optimal solution for cost-constrained research initiatives, particularly under these conditions:
- Availability of legacy SWATH libraries (minimizing database generation expenses);
- Requirement for exceptional instrument reliability (annual operational failures < 0.5);
- Need for label-free flux quantification during ongoing studies (throughput: ~30 samples weekly).
- Its capabilities are maximized through an integrated workflow combining dynamic SWATH windowing, online enrichment, and cloud-based data processing. When processing sample quantities exceeding 10μg, this platform delivers superior cost-effectiveness relative to higher-end instrumentation.
(D) Astral Mass Spectrometer (Thermo Fisher Scientific)
Core Advantages
- Unprecedented Performance: Preliminary data indicates ~10× faster acquisition and 10× sensitivity improvements versus current platforms.
- Innovative Analyzer Design: Novel architecture promises transformative gains in analytical depth and speed.
Current Status
- Recently launched platform with developing application case studies
- Phosphoproteomics best practices still evolving
- Long-term performance validation and reliability assessment ongoing
Potential Applications
- Future single-cell phosphoproteomics research
- Ultra-high-throughput proteomic screening
- Applications demanding extreme sensitivity and rapid analysis cycles
Platform Selection Decision Pathway: Practical Implementation Guide
- Decision Node 1: Quantification Precision Requirements
- Is maximum quantification accuracy (particularly for TMT experiments) the project's primary objective?
- Yes: Orbitrap Eclipse is recommended due to MS3 quantification capability
- No: Proceed to Node 2
- Decision Node 2: Throughput and Scale Demands
- Does the study involve large sample cohorts (>100 cases) or require minimal turnaround time?
- Yes: timsTOF Pro/HT systems are preferred for PASEF/diaPASEF high-throughput operation
- No: Advance to Node 3
- Decision Node 3: Sample Quantity Limitations
- Is biological material extremely limited (e.g., single-cell or biopsy specimens)?
- Yes: Prioritize Orbitrap Eclipse or timsTOF SCP for optimal sensitivity
- No: Continue to Node 4
- Is biological material extremely limited (e.g., single-cell or biopsy specimens)?
- Decision Node 4: Acquisition Strategy and Budget
- Will data-independent acquisition (DIA) be implemented with moderate budget constraints?
- Yes: Consider either:
- a) TripleTOF 7600 (established SWATH® methodology)
- b) timsTOF (diaPASEF implementation)
- No: Proceed to Node 5
- Decision Node 5: Advanced Technical Specifications
- Are maximum mass resolution or electron-transfer dissociation (ETD) fragmentation essential?
- Yes: Orbitrap platforms are optimal
- No: Conduct comprehensive evaluation of:
- Budgetary parameters
- Existing instrumentation access
- Technical preferences
- (Note: timsTOF excels in throughput, Orbitrap delivers superior overall performance, TripleTOF offers exceptional cost efficiency)
Select Service
Learn more
In-Depth Analysis of Key Technical Parameters
1. Sensitivity and Limit of Detection (LOD) Benchmark Comparison
| Platform | Phosphopeptide LOD (HeLa) | Key Influencing Factors | Recommended Applications |
|---|---|---|---|
| Orbitrap Eclipse | 10-50 amol (MS1) | Ion transmission efficiency, Trap charge capacity | Single-cell / Trace tissue (<1,000 cells) |
| timsTOF SCP | 5-20 amol (diaPASEF) | TIMS focusing effect, Quadrupole transmission | Needle biopsy specimens, Organoid micro-regions |
| TripleTOF 7600 | 100-500 amol (SWATH) | Quadrupole transmission losses | Conventional samples (>10 μg protein) |
- Note: Actual LOD requires comprehensive evaluation considering chromatographic systems (nanoLC vs. microLC) and column loading capacity.
2. Core Quantitative Precision Metrics
| Parameter | Orbitrap (MS3-TMT) | timsTOF (MS2-TMT) | TripleTOF (MS2-TMT) |
|---|---|---|---|
| CV (10plex) | < 5% | 8-15% | 10-18% |
| Interference Factor (IF) | 0.05-0.1 | 0.2-0.5 | 0.3-0.6 |
| Optimal Use Case | Pharmacodynamic studies | High-throughput screening | Cost-effective multiplexed quantitation |
- Definition: Interference Factor (IF) = Signal leakage rate from adjacent reporter ion channels. Lower values indicate reduced co-isolation interference.
Platform-Specific Solutions for Complex Scenarios
1. Phosphotyrosine (pY) Enrichment Detection
Challenge: pY accounts for merely 0.05% of all phosphorylation events, with conventional enrichment exhibiting low efficiency.
Optimized Workflow:
- Platform: Orbitrap Eclipse (leveraging pY antibody pre-enrichment + ultrahigh sensitivity)
- Performance Gain: Achieves 50-fold enhancement in pY site identification versus standard workflows.
2. Multi-phosphopeptide Resolution
- Challenge: Incomplete fragmentation of high-charge-state peptides leads to ambiguous site localization.
- Platform Comparison:
| Parameter | Orbitrap (EThcD) | timsTOF (PASEF-CID) | TripleTOF (CID-HCD) |
|---|---|---|---|
| Localization Rate | >85% (4+ charge state) | 60-70% | 50-65% |
| Spectral Quality | Backbone ions + modification retention | Partial backbone ion loss | Neutral loss dominance |
Cost-Benefit Modeling Analysis (100-Sample Project)
| Cost Component | Orbitrap HF-X | timsTOF Pro 2 | TripleTOF 7600 |
|---|---|---|---|
| Instrument Depreciation | $300/sample | $200/sample | $150/sample |
| Consumables (Columns/Reagents) | $80/sample | $70/sample | $60/sample |
| Bioinformatics Labor | $200/sample (DIA) | $150/sample (diaPASEF) | $180/sample (SWATH) |
| Total Cost | $580/sample | $420/sample | $390/sample |
| Value Proposition | Ultrahigh-precision quantitation | Depth-throughput balance | Stable batch processing |
To learn more about the workflow, please refer to "Phosphoproteomics Workflow Explained: From Sample to Data".
How to choose between DIA and DDA, please refer to "DIA vs DDA in Phosphoproteomics: Which One Should You Use?".
People Also Ask
What mass spectrometer is most suitable for targeted proteomics quantification?
Although essentially any tandem MS instrument can be used for targeted quantitation, triple quadrupole and quadrupole-ion trap (Q-Trap) MS instruments are most widely used.
What are the techniques used in phosphoproteomics?
Typical phosphoproteomics workflows involve sample enrichment followed by mass spectrometry (MS) analysis using complementary fragmentation techniques (CID, HCD, EThcD, and ETD).
What is quantitative phosphoproteomics?
LC-MS/MS based quantitative phosphoproteomics provides a powerful tool to analyze site-specific protein phosphorylation.
What is phosphoproteomics mass spectrometry?
Proteins for phosphoproteomic analysis come from a variety of sources and must first be extracted and denatured, then reduced, alkylated and digested into peptides. Phosphopeptides are then enriched before they can be analyzed by LC-MS/MS on Orbitrap-based mass spectrometers.
References
- Mathew A, Giskes F, Lekkas A, Greisch JF, Eijkel GB, Anthony IGM, Fort K, Heck AJR, Papanastasiou D, Makarov AA, Ellis SR, Heeren RMA. "An Orbitrap/Time-of-Flight Mass Spectrometer for Photofragment Ion Imaging and High-Resolution Mass Analysis of Native Macromolecular Assemblies." J Am Soc Mass Spectrom. 2023 Jul 5;34(7):1359-1371. doi: 10.1021/jasms.3c00053
- Lou R, Cao Y, Li S, Lang X, Li Y, Zhang Y, Shui W. "Benchmarking commonly used software suites and analysis workflows for DIA proteomics and phosphoproteomics." Nat Commun. 2023 Jan 6;14(1):94. doi: 10.1038/s41467-022-35740-1
- Paulo JA, Schweppe DK. "Advances in quantitative high-throughput phosphoproteomics with sample multiplexing." Proteomics. 2021 May;21(9):e2000140. doi: 10.1002/pmic.202000140
- Lancaster NM, Sinitcyn P, Forny P, Peters-Clarke TM, Fecher C, Smith AJ, Shishkova E, Arrey TN, Pashkova A, Robinson ML, Arp N, Fan J, Hansen J, Galmozzi A, Serrano LR, Rojas J, Gasch AP, Westphall MS, Stewart H, Hock C, Damoc E, Pagliarini DJ, Zabrouskov V, Coon JJ. "Fast and deep phosphoproteome analysis with the Orbitrap Astral mass spectrometer." Nat Commun. 2024 Aug 15;15(1):7016. doi: 10.1038/s41467-024-51274-0
- Andrews GL, Simons BL, Young JB, Hawkridge AM, Muddiman DC. "Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600)." Anal Chem. 2011 Jul 1;83(13):5442-6. doi: 10.1021/ac200812d
- Wörner TP, Aizikov K, Snijder J, Fort KL, Makarov AA, Heck AJR. "Frequency chasing of individual megadalton ions in an Orbitrap analyser improves precision of analysis in single-molecule mass spectrometry." Nat Chem. 2022 May;14(5):515-522. doi: 10.1038/s41557-022-00897-1











