Total TRANS-Fatty Acid Analysis Service
Trace-level trans-fatty acid (TFA) detection is no longer optional — it's essential for compliant labeling, product benchmarking, and source attribution. Yet most labs still struggle with cis/trans overlap, isomer resolution, and matrix interference.
Creative Proteomics delivers high-resolution total trans-fatty acid quantification and isomer profiling using validated GC–FID workflows, GC–MS identity confirmation, and optional silver-ion HPLC. Whether you're verifying label claims, tracing industrial vs ruminant origins, or optimizing formulations, our methods give you the confidence to report, compare, and decide.
We Help You Solve:
- Ambiguous cis/trans overlap that inflates or deflates trans-fat totals
- Positional isomer interference, especially across C18:1 trans series
- Matrix suppression in complex samples (oils, processed foods, biofluids)
- Label risk in noncompliant ΣTFA reporting or unclear source attribution
- Inconsistent derivatization that impacts yield and reproducibility
Submit Your Request Now
×
- What We Provide
- Advantages
- Technology Platform
- Sample Requirements
- Demo
- FAQs
What Are Total Trans-Fatty Acids?
Trans-fatty acids are unsaturated fatty acids containing at least one trans double bond. They arise from industrial hydrogenation and ruminant biohydrogenation pathways.
Total trans-fatty acid analysis sums all trans isomers after resolving cis/trans and positional variants. The approach enables accurate labeling checks, risk assessment, and R&D comparisons across foods, oils, and biofluids.
What Problems Does This Service Solve?
- Ambiguous cis/trans separation. We resolve coeluting peaks that inflate or deflate totals.
- Positional isomer overlap. Trans fat profiling distinguishes C18:1t subforms for clear attribution.
- Matrix interference. Methods are tuned for oils, processed foods, dairy, and biofluids.
- Derivatization variability. Controlled FAME preparation ensures consistent conversion and yield.
- Label-claim verification. Total trans-fatty acid analysis supports compliant, defensible reporting.
- Source differentiation. Patterns separate industrial hydrogenation from ruminant origins.
Service Scope: Total Trans-Fatty Acid Quantification & Profiling
- Quantitative Analysis of Total Trans-Fatty Acids. GC-FID primary quantitation with GC-MS support across foods, oils, tissues, and cell samples.
- Isomer-Specific Profiling. Separate and quantify individual trans isomers, including trans-oleic and trans-linoleic species.
- Cis/Trans Ratio Assessment. Evaluate cis/trans ratios to gauge lipid integrity and processing effects.
- FAME Derivatization. Standardized methylation workflow enables precise, reproducible chromatographic separation.
- Source Fingerprinting. Interpret industrial versus ruminant signatures using diagnostic isomer patterns.
- Orthogonal Identity Confirmation. Resolve ambiguous peaks via GC-MS; optional silver-ion HPLC for challenging pairs.
- Custom Reporting Panels. Configure summations, ratios, and units to match SOPs and market needs.
Analyte Coverage: Detectable Trans-Fatty Acid Isomers & Ratios
Detectable Isomers by Category
| Category | Examples / Isomers | Reporting Notes |
|---|---|---|
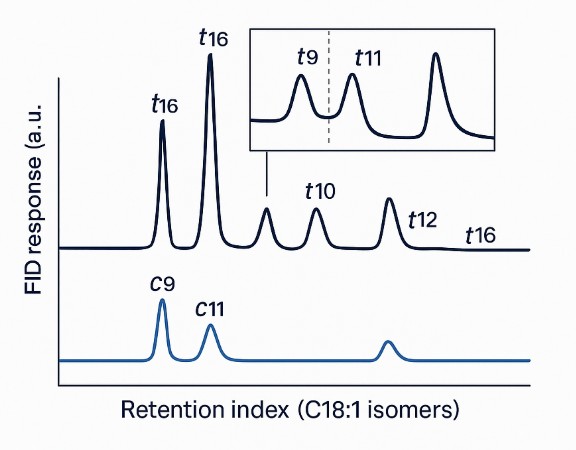
| Trans MUFA | C16:1 trans (palmitelaidic); C18:1 trans series t6–t16 (incl. elaidic t9, vaccenic t11); C20:1 trans; C22:1 trans | Reported as individual isomers when resolved; low-abundance long-chain species reported when signal quality permits |
| Trans PUFA (non-conjugated) | C18:2 trans sets (tt, tc, ct; e.g., linoelaidic); C18:3 trans-containing isomers (ALA-derived) | Quantified as resolved isomers or grouped sets depending on chromatographic separation |
| Conjugated systems (optional) | CLA isomers (e.g., c9,t11; t10,c12) | Profiled separately and distinguished from non-conjugated trans species |
Configurable Totals & Indices
| Metric | Description | Typical Use |
|---|---|---|
| Σ TFA (total trans) | Sum of all detected trans-fatty acids | Label verification; trend tracking |
| Σ C18:1t / Σ C18:2t / Σ C18:3t | Class-level trans summaries by carbon chain and unsaturation | Method comparison; matrix benchmarking |
| Cis/trans ratios | Ratio of cis vs trans within a class or total pool | Integrity assessment; processing impact |
| Diagnostic ratios | Isomer patterns supporting industrial vs ruminant source interpretation | Source fingerprinting and risk assessment |
Final reporting is matrix-dependent and reflects column chemistry and any orthogonal confirmation used (GC-FID, GC-MS, optional silver-ion HPLC).
Why Choose Our Total Trans-Fatty Acid Analysis
- Baseline cis/trans resolution: Highly polar long-column GC (SP-2560/CP-Sil 88 class) achieves Rs ≥ 1.5 for key C18:1 trans pairs under validated conditions.
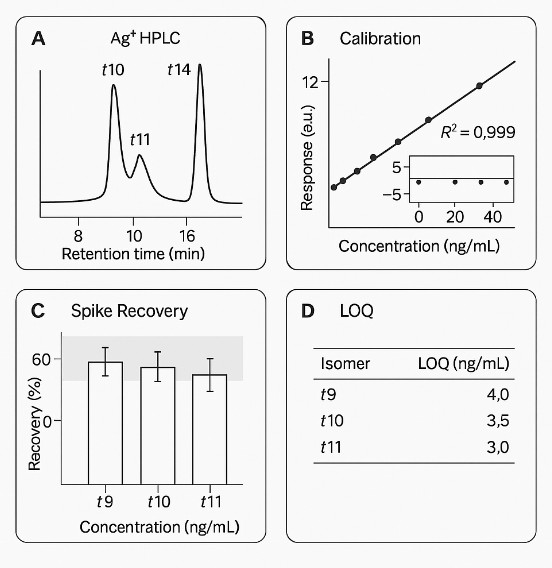
- Quantitative confidence: Multi-point calibration with internal standards; typical study metrics include R2 ≥ 0.995, spike recovery ~85–115%, and repeatability RSD ≤ 10% (matrix-dependent; reported per project).
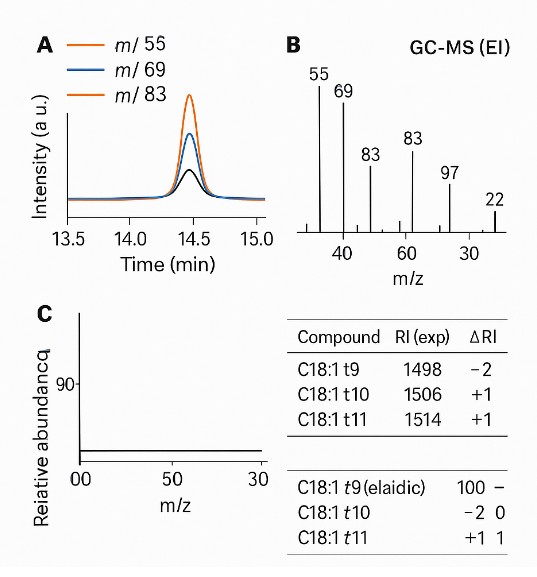
- Orthogonal identity checks: GC–MS (EI) spectral confirmation with retention-index matching; optional silver-ion HPLC for challenging positional isomers (e.g., t10/t11).
- Matrix-aware methods: Optimized prep for oils, processed foods, dairy, meats, feeds, plasma/serum, and tissues to minimize matrix suppression and carryover.
- Decision-ready outputs: ΣTFA, cis/trans ratios, diagnostic isomer ratios, QC dashboards, chromatograms, and machine-readable peak tables for rapid internal review.
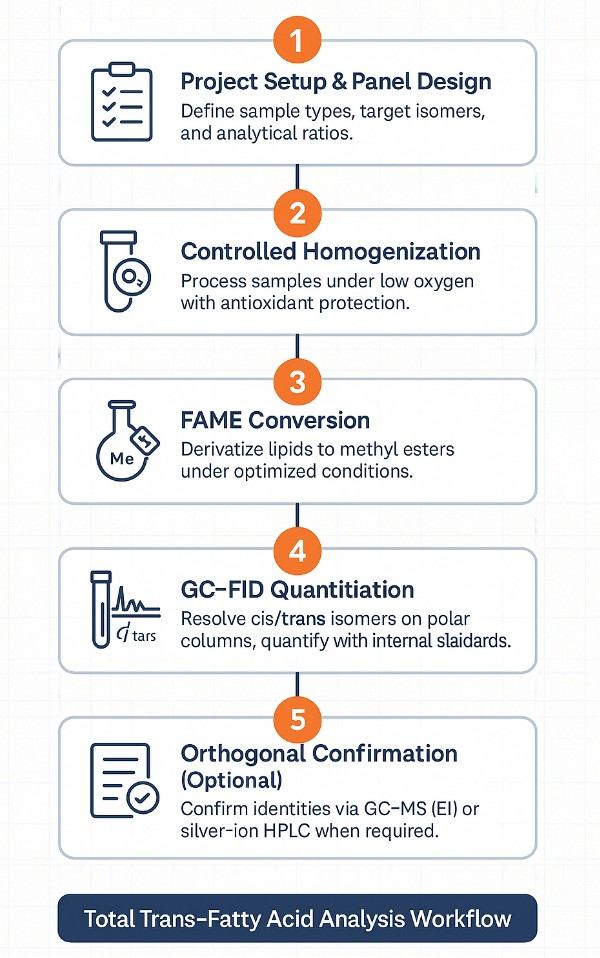
Total Trans-Fatty Acid Analysis Workflow: Step-by-Step Process
- Intake & Panel Design
Define matrices, reporting units, target isomers, and any diagnostic ratios. - Sample Homogenization & Controls
Homogenize under low-oxygen conditions; apply antioxidant controls when matrix warrants. - FAME Derivatization
Convert lipids to methyl esters under controlled conditions to ensure consistent yields. - GC-FID Separation & Quantitation
Resolve cis/trans and positional isomers on highly polar columns; quantify with internal standards. - Orthogonal Confirmation (As Needed)
Use GC–MS (EI) for identity checks; optional silver-ion HPLC for challenging pairs. - Quantification & QC Review
Apply multi-point calibration, blanks, duplicates, and spike recoveries; verify data integrity. - Reporting & Interpretation
Deliver ΣTFA, isomer tables, cis/trans ratios, diagnostic indices, QC dashboards, and chromatograms.
Total Trans-Fatty Acid Instrumentation & Platform Capabilities
GC–FID (primary quantitation): Highly polar long columns (SP-2560 / CP-Sil 88 class) for cis/trans resolution.
GC–MS (EI) confirmation: Spectral libraries and retention-index matching to verify ambiguous or trace peaks.
Silver-ion HPLC (optional): Orthogonal separation for challenging positional or geometric isomer pairs.

1260 Infinity II HPLC (Figure from Agilent)

Agilent 7890B-5977A (Figure from Agilent)
How to Submit Samples for Total TFA Testing
| Matrix / Sample Type | Preferred Form | Minimum Amount (per sample) | Backup Aliquot | Notes |
|---|---|---|---|---|
| Edible oils, fats | Neat liquid/solid | 0.5–1 g | 1 × 0.5 g | Avoid antioxidants unless part of product; cap tightly to limit oxygen. |
| Processed foods, snacks, bakery | Homogenized solid | 2–5 g | 1 × 2 g | Provide full ingredient list if available; homogenize thoroughly to reduce heterogeneity. |
| Dairy (butter, cheese, milk powder) | Homogenized solid/powder | 2–5 g | 1 × 2 g | Keep cold; minimize light exposure to reduce oxidation. |
| Meats, meat products | Homogenized tissue | 2–5 g | 1 × 2 g | Remove excess bone/casing; record fat content if known. |
| Animal feeds | Homogenized solid | 2–5 g | 1 × 2 g | Grind to uniform particle size if permissible. |
| Serum / plasma | Liquid, frozen | 100–200 µL | 1 × 100 µL | EDTA or heparin acceptable; avoid hemolysis; no preservatives unless disclosed. |
| Cell pellets | Frozen pellet | ≥5 × 106 cells | 1 matching pellet | Wash with PBS, remove media; record cell line and culture conditions. |
| Tissue | Frozen tissue | 50–100 mg | 1 × 50 mg | Snap-freeze promptly; avoid repeated freeze–thaw cycles. |
Demo Results
FAQ of TRANS-Fatty Acid Analysis Service
How do you handle matrices with very low fat content or unusual lipids?
We customise sample preparation protocols when fat levels are minimal or lipid profiles atypical, adjusting internal‑standard loads and extraction steps to ensure quantifiable recovery of trans isomers from challenging matrices.
Can you detect trace trans‑isomers below regulatory thresholds?
Yes — our method sensitivity supports reporting well below typical regulatory cut‑offs, using low‑level calibration, rigorous spike recovery and orthogonal confirmation where needed to validate trace‑level peaks.
How do you ensure the identity of co‑eluting or ambiguous peaks in complex foods?
We employ orthogonal techniques including EI mass spectral library matching, retention‑index verification and optionally silver‑ion HPLC separation to confirm challenging positional/geometric isomer pairs and remove ambiguity.
Are the diagnostic source‑fingerprinting ratios based solely on industrial vs ruminant origins?
No — while industrial versus ruminant signatures (e.g., t9/t11 patterns) are a key component, our interpretation also accounts for matrix context, production process and compositional background, enabling informed attribution rather than simple classification.
What level of reporting detail can you provide in your deliverables?
Our reports include full isomer‑specific tables, cis/trans ratios, ΣTFA summaries, chromatograms, calibration/PUQC data and machine‑readable peak lists; they are configured to align with client SOPs or market‑specific reporting frameworks.
Will method performance vary across food, oil, dairy and biofluid matrices?
Performance parameters such as signal‑to‑noise, spike recovery and repeatability do depend on matrix complexity; we document matrix‑specific QC metrics per job and provide context for any deviations rather than relying on a one‑size‑fits‑all performance claim.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the metabolomics-related papers published by our clients:

- Proteolytic activation of fatty acid synthase signals pan-stress resolution. Nature Metabolism.
- Prospective randomized, double-blind, placebo-controlled study of a standardized oral pomegranate extract on the gut microbiome and short-chain fatty acids. Foods.
- Calcium homeostasis and stable fatty acid composition underpin heatwave tolerance of the keystone polychaete Hediste diversicolor. Environmental Research.
- B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nature communications.