DNA Pull-Down Assay Service | DNA–Protein Interaction
Understanding how transcription factors and regulators bind to DNA is essential to decode gene expression and pathway control. Creative Proteomics’s DNA Pull-Down Assay Service helps researchers identify and validate DNA-binding proteins with precision — from targeted confirmation to large-scale discovery.
By combining biotin/desthiobiotin probe chemistry, streptavidin magnetic capture, and LC–MS/MS or Western blot readouts, we deliver reproducible data that drives publications and mechanistic insight.
Why researchers choose Creative Proteomics
- Comprehensive DNA–Protein Interaction Analysis — from hypothesis testing to proteomic discovery.
- Ultra-Sensitive Detection — enrich low-abundance DNA-binding proteins (<1% nuclear protein).
- Optimized Stringency — customized buffers and controls reduce background, improving signal-to-noise ≥10×.
- Bioinformatics Integration — GO, KEGG, and STRING analyses included for meaningful interpretation.
Start your DNA pull-down project today and discover the proteins driving your gene regulation.
Submit Your Request Now
×
- What We Provide
- Method Comparison
- Advantages
- Workflow
- Sample Requirements
- Demo
- Use Cases
- FAQs
What Is a DNA Pull-Down Assay?
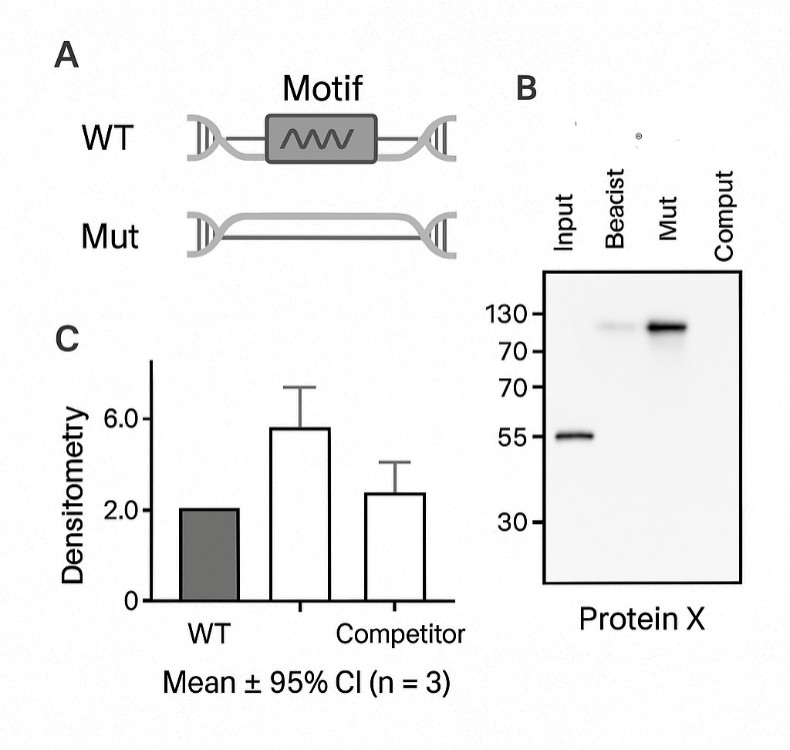
DNA pull-down is an in-vitro method to study DNA–protein interactions. A biotin or desthiobiotin–labeled double-stranded DNA probe representing a promoter, enhancer, or variant-altered motif is immobilized on streptavidin magnetic beads. The probe–bead complex is incubated with a nuclear extract, washed under nuclease-free conditions, and eluted. Captured proteins are then analyzed by Western blot for targeted validation or LC–MS/MS for unbiased discovery.
Key strengths
- Sensitive enrichment of low-abundance DNA-binding proteins
- Flexible probe design (5′-biotin oligos or PCR-labeled probes)
- Direct compatibility with downstream proteomics workflows
Considerations
- Long probes can raise non-specific background; include mutant or competitor controls
- Strict nuclease-free handling is essential throughout the workflow
- As an in-vitro assay, results should be complemented by in-cell methods (e.g., ChIP-qPCR)
DNA Pull-Down Assay Options — Western Blot Validation or LC–MS/MS Discovery
DNA pull-down — Western blot (validation)
This path confirms whether a named candidate, such as a transcription factor, binds a defined promoter or enhancer sequence. It uses a biotin or desthiobiotin–labeled double-stranded DNA probe immobilized on streptavidin magnetic beads, followed by Western blot detection with client-supplied or sourced antibodies.
What's included
- Probe design and synthesis (5′-biotin oligos, PCR incorporation such as Biotin-11-dUTP, or biotin-TEG spacers)
- Streptavidin bead coupling, incubation with nuclear extract, and optimized washes
- Elution of DNA–protein complexes under conditions aligned to the probe chemistry
- Western blot for specified targets, with appropriate positive/negative controls
- QC images and a structured report covering methods, parameters, and interpretation
When to choose this option
- You have a hypothesis-driven target (e.g., predicted TF–promoter interaction)
- You need orthogonal confirmation for figures or follow-up functional assays
DNA pull-down — LC–MS/MS (discovery)
This path profiles proteins that interact with the DNA region of interest without pre-selecting targets. Pull-down eluates are resolved and identified by mass spectrometry, then summarized with bioinformatics.
What's included
- Probe design and synthesis with wild-type plus optional mutant or competitor control
- Pull-down with stringent wash strategy and probe/eluate QC (e.g., electrophoresis shift, silver stain)
- LC–MS/MS identification of proteins from eluates
- Candidate interactor table highlighting group-unique or prioritized proteins
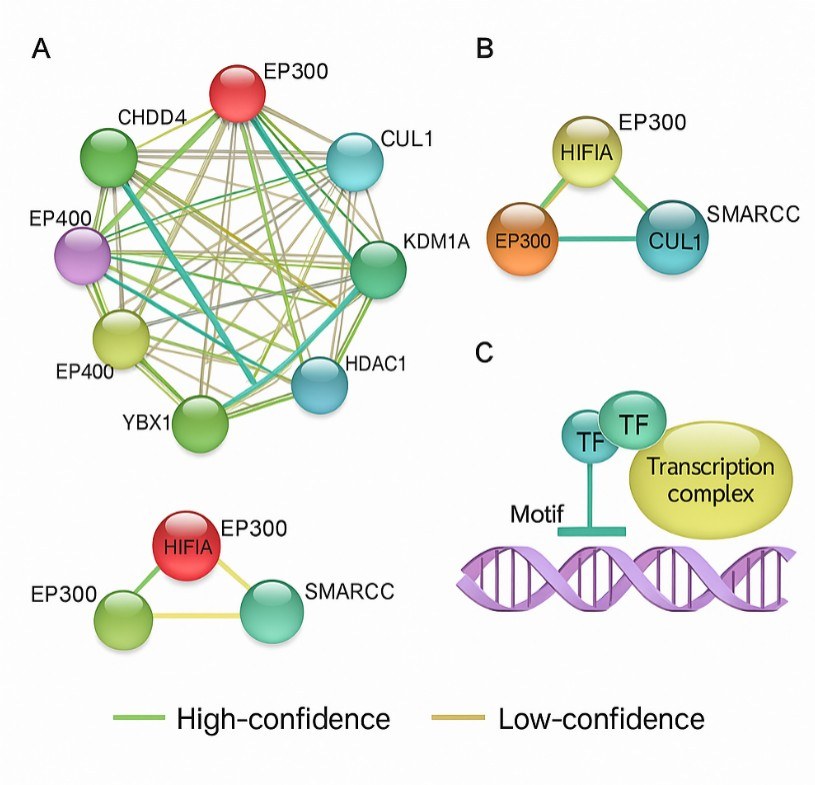
- Bioinformatics summaries: GO terms, KEGG pathways, and STRING protein–protein networks
- Comprehensive report with decision guidance for downstream validation
When to choose this option
- You aim to discover unknown binders at a promoter, enhancer, or variant-altered motif
- You need a prioritized short list to guide Western blot or functional follow-ups
Quality gate and acceptance criteria
Assay success is assessed against defined controls. The positive control should detect or identify the expected binder, and the negative control should not. Any exceptions are documented with technical rationale and proposed next steps.
Probe Labeling Options — Which Handle Should I Choose?
Selecting the right affinity handle affects elution strategy and background. Probes can be prepared as terminally biotinylated oligos, via PCR incorporation (for example, Biotin-11-dUTP), or with biotin-TEG spacers. Desthiobiotin is available for reversible capture and gentle elution.
| Label | Coupling to streptavidin | Elution behavior | When to use |
|---|---|---|---|
| Biotin | Very high affinity | Denaturing elution or on-bead digestion; strong retention during stringent washes | Routine pull-downs requiring high stringency |
| Desthiobiotin | High-affinity but reversible | Gentle competitive elution with biotin; helps preserve complex integrity | When native-like recovery or mild conditions are preferred |
Note: Biotin-14-CTP is used to label RNA during in-vitro transcription and applies to RNA pull-down, not DNA pull-down. Request RNA pull-down if your objective is RNA–protein capture.
Method Comparison: DNA Pull-Down vs ChIP-qPCR vs EMSA vs Yeast One-Hybrid
| Criterion | DNA Pull-Down | ChIP-qPCR | EMSA / Gel-Shift | Yeast One-Hybrid (Y1H) |
|---|---|---|---|---|
| Primary question | Which proteins bind a defined DNA sequence (discovery or validation)? | Does a known protein occupy a specific genomic locus in cells? | Does a protein bind a short motif in vitro (binding yes/no, affinity/shift)? | Can a TF bind/activate a reporter via the test DNA in yeast? |
| Readout | LC–MS/MS (unbiased IDs) and/or Western blot (targeted) | qPCR enrichment at primered region | Mobility shift; supershift with antibody | Reporter activation/colony growth |
| Biological context | In-vitro enrichment using biotin/desthiobiotin probes | In-cell (crosslinked or native chromatin) | In-vitro (purified protein or extract) | Heterologous yeast system |
| Discovery vs validation | Both (MS = discovery; WB = validation) | Validation only at known loci | Validation/affinity for short motifs; no discovery | Screening TF–DNA interactions via reporter |
| Key inputs | DNA probe; antibody only if using WB | High-quality antibody for target protein | DNA oligo probe; purified protein or nuclear extract; antibody optional for supershift | DNA bait cloned to reporter; yeast host strain |
| Specificity controls | WT vs mutant/competitor probes; beads-only negative | IgG/no-Ab; off-target loci | Competitor/mutant oligos; cold competitor; supershift | Empty vector; non-specific reporter controls |
| Sensitivity to low-abundance targets | High (enrichment + MS depth) | Variable (antibody/biomaterial dependent) | Moderate; gel detection limits apply | Variable; depends on expression in yeast |
| What you learn | Protein identities binding your sequence; confirm candidates | Whether a specific protein binds a locus in cells | Whether a protein binds a short DNA motif and relative affinity | Whether a TF can drive binding/activation via the sequence |
| Best used when | You need IDs or orthogonal confirmation for promoter/enhancer studies | You need in-cell occupancy evidence for a known factor | You need a quick motif-level binding check or Kd estimate | You need TF screening or motif activity readout in a reporter system |
| Key limitations | In-vitro context; long probes can raise background (mitigate with controls) | Antibody quality and crosslink bias; locus-specific | No protein ID without antibody; potential gel artifacts | Yeast context may not mirror native regulation |
Why Choose Creative Proteomics for DNA Pull-Down Assays
- High sensitivity for low-abundance proteins: Enrichment protocol detects DNA-binding proteins at<1% nuclear abundance with consistent recovery.
- Flexible probe design: Supports wild-type, mutant, and competitor sequences up to 100+ bp for precise interaction profiling.
- Optimized stringency: Wash conditions tuned per project to achieve ≥10× signal-to-noise improvement over generic protocols.
- Integrated proteomics and analysis: LC–MS/MS includes peptide-level FDR ≤1% and functional annotation via GO, KEGG, and STRING.
- Sample-efficient workflows: Accepts limited material without compromising data quality—ideal for scarce or valuable samples.
- Ready for downstream applications: Clean outputs compatible with EMSA, ChIP-qPCR, pathway mapping, and network modeling.
Workflow for DNA Pull-Down Analysis
- Project scoping
Define research goals, target DNA sequence or motif, control design, and detection method (Western blot or LC–MS/MS). - Template preparation (if required)
Prepare DNA template via T-cloning or PCR to support probe synthesis. - Probe design & labeling
Generate biotin- or desthiobiotin–labeled double-stranded DNA probes (wild-type ± mutant or competitor sequences). - Bead coupling
Attach labeled probes to streptavidin magnetic beads under controlled conditions. - Incubation with nuclear extract
Expose bead–probe complexes to nuclear protein under nuclease-free conditions to capture specific DNA–protein interactions. - Stringent washes
Remove unbound and non-specifically bound proteins while preserving interaction fidelity. - Elution
Release DNA-bound proteins using reversible elution (desthiobiotin) or denaturing/on-bead strategies (biotin). - Detection & identification
Analyze eluates by Western blot for known targets and/or LC–MS/MS for unbiased protein discovery.


Q Exactive HF-X MS (Figure from Thermo)

SCIEX Triple Quad™ 6500+ (Figure from Sciex)
Sample Requirements for DNA Pull-Down Assay Analysis
To ensure high assay quality and reproducibility, please prepare and submit samples according to the specifications below. We accept both fresh biological materials and pre-extracted nuclear proteins. If unsure, consult your project coordinator before shipment.
| Sample Type | Recommended Amount per Group | Concentration & Purity | Storage & Shipping | Notes |
|---|---|---|---|---|
| Animal Cells (e.g., HEK293, HeLa) | WB: ≥3 × 107 cells MS: ≥5 × 107 cells | N/A for whole cells Nuclear extract: ≥300 µg total protein, ≥0.5 mg/mL | Snap-freeze in liquid nitrogen (open cap); store at −80 °C; ship on dry ice | Use PBS wash to remove media/serum. Trypsinize adherent cells gently. Avoid freeze–thaw cycles. |
| Animal Tissue (e.g., liver, brain) | WB: ≥1 g MS: ≥2 g | N/A | Chop to ~200 mg pieces; snap-freeze in open tube; store at −80 °C; ship on dry ice | Minimize tissue handling time (<1 min). Avoid RNase/DNase contamination. |
| Plant Leaf Tissue | WB: ≥2 g MS: ≥4 g | N/A | Same as above | Use young or fresh leaves when possible. Avoid yellowing or wilted samples. |
| Plant Root or Fruit | WB: ≥4 g MS: ≥8 g | N/A | Same as above | Ensure root samples are free of soil; fruit should be deseeded when applicable. |
| Microbial Pellets (e.g., yeast, E. coli) | WB: ≥50 μL packed cells MS: ≥100 μL packed cells | OD600 ≥ 2.0 (for culture) | Pellet, remove supernatant, snap-freeze, and store at −80 °C; ship on dry ice | Provide strain name and OD measurement. |
| Nuclear Extracts | ≥100 µg per group; ≥300 µg total | >0.5 mg/mL; buffer must be nuclease-free and clearly labeled | Aliquot in low-retention tubes; store at −80 °C; ship on dry ice | Include composition, protease inhibitors used, and preparation method. |
| Plasmid DNA (for probe template) | ≥4 µg; ≥200 ng/µL | OD260/280 = 1.6–2.0 | Store at −20 °C; ship with ice packs | Provide vector map, resistance marker, and sequencing verification. |
| PCR Products (optional) | ≥500 ng | Purified; gel-checked | Store at −20 °C; ship with ice packs | Can be used for direct probe labeling (e.g., Biotin-11-dUTP). |
Demo Results
Use Cases for DNA Pull-Down Assays
Promoter–Transcription Factor Validation
Confirm whether specific transcription factors bind to promoter regions using motif predictions, prior datasets, or literature references.
Enhancer and Silencer Interaction Profiling
Identify proteins that interact with distal regulatory elements, such as enhancers or silencers, to better understand transcriptional regulation.
Comparative Binding Analysis
Compare DNA–protein interaction profiles across different conditions, treatments, genotypes, or time points to detect regulatory shifts.
Pre-screening for ChIP-Based Assays
Use pull-down results to narrow down candidate binders before committing to more resource-intensive in-cell assays like ChIP-qPCR.
Pathway and Network Mapping
Generate candidate protein lists for downstream bioinformatics analysis, including pathway enrichment, interaction networks, and functional clustering.
Species-Specific DNA–Protein Studies
Apply custom-designed probes to study DNA-binding regulators in plant or microbial systems, where cross-reactivity or antibody availability is limited.
FAQ of DNA Pull-Down Assay
Is a DNA pull-down assay in vitro or in cellulo, and what does that mean for interpretation?
DNA pull-down is an in-vitro capture method: a labeled double-stranded DNA fragment is immobilized on streptavidin beads and incubated with protein extracts to enrich sequence-specific binders; results reflect biochemical specificity under controlled conditions and should be complemented with in-cell assays (e.g., ChIP-qPCR) when locus occupancy in living cells is required.
When should I choose DNA pull-down instead of ChIP-qPCR?
Use DNA pull-down when you need the identity of proteins that bind a defined sequence (discovery by LC–MS/MS, or validation by Western blot); choose ChIP-qPCR when you already have a specific protein in mind and need evidence of its occupancy at a genomic locus inside cells.
What controls help distinguish specific from non-specific binding?
Pair the wild-type probe with a motif-disrupted mutant or competitor probe, include beads-only or unrelated-sequence controls, and document wash conditions; this combination reduces background and increases confidence that enrichment reflects motif-dependent binding.
Biotin vs desthiobiotin: which probe handle should I use?
Biotin–streptavidin binding is extremely tight and tolerates harsh washes, suitable for stringent enrichment; desthiobiotin is a biotin analog with lower affinity that enables gentle competitive elution with free biotin, useful when you want reversible recovery of bound proteins or milder conditions.
Do I need antibodies for a DNA pull-down experiment?
Antibodies are only required when the readout is Western blot against named candidates; for LC–MS/MS discovery, identification relies on mass spectrometry and database searches rather than target-specific antibodies.
Should I send nuclear extracts or whole-cell lysates?
Nuclear extracts generally improve signal-to-noise for transcription-related binders because many DNA-binding regulators are concentrated in the nucleus; whole-cell lysates can be considered for targets expected outside the nucleus, but may increase non-specific background.
How do probe design choices affect specificity and background?
Probes that focus on the functional motif and avoid repetitive/high-GC regions typically reduce non-specific interactions; pairing a wild-type probe with a sequence-altered control allows direct attribution of binding to the intended motif and supports confident downstream validation.
What kind of data will I receive from LC–MS/MS discovery?
You receive QC evidence (e.g., silver-stained gels), protein identification tables with false-discovery-rate control, and functional context such as GO/KEGG pathway and STRING network summaries, enabling rapid prioritization of candidate interactors for follow-up tests.
Can DNA pull-down replace yeast one-hybrid (Y1H)?
Pull-down can bypass library construction and directly enrich native nuclear proteins that bind your sequence, providing a proteomics-grade candidate list; however, Y1H reports transcriptional activation potential in a reporter context, so it remains complementary when you need activity readouts rather than biochemical binding alone.
Is RNA pull-down the same as DNA pull-down?
No; RNA pull-down uses biotinylated RNA baits transcribed in vitro to capture RNA-binding proteins, often revealing different interaction patterns due to RNA secondary structure, while DNA pull-down targets DNA-binding proteins recognizing double-stranded DNA motifs.
Learn about other Q&A about proteomics technology.
Publications
Here are some publications in Proteomics research from our clients:

- Hyperfunction of post-synaptic density protein 95 promotes seizure response in early-stage aβ pathology. 2024. https://doi.org/10.1038/s44319-024-00090-0
- Quantitative Proteomic Analysis of Cellular Responses to a Designed Amino Acid Feed in a Monoclonal Antibody Producing Chinese Hamster Ovary Cell Line. 2018. https://doi.org/10.29252%2F.22.6.385
- In vitro assays reveal inherently insecticide-tolerant termite symbionts. 2023. https://doi.org/10.3389/fphys.2023.1134936
- Epithelial WNT2B and Desert Hedgehog are necessary for human colonoid regeneration after bacterial cytotoxin injury. 2018. https://doi.org/10.1101/434639
- Confirmation of translatability and functionality certifies the dual endothelin1/VEGFsp receptor (DEspR) protein. 2016. https://doi.org/10.1186/s12867-016-0066-8