Drug Affinity Responsive Target Stability (DARTS) Assay Service
Accurately Decipher Protein–Small Molecule Interactions to Accelerate Drug Discovery
Creative Proteomics offers a comprehensive DARTS assay platform to help you:
- Validate Mechanisms of Action (MOA)
- Confirm Target Engagement
- Identify Off-Target Effects
- Rank Binding Affinities across Analogs
Our label-free, physiologically relevant, and mass spectrometry-driven approach ensures confident, high-resolution drug–protein interaction profiling across diverse small molecules and protein classes.
Submit Your Request Now
×
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQ
Understand Drug–Protein Interactions with Precision
In modern drug development, identifying direct and specific interactions between small molecules and their protein targets is fundamental to discovering mechanisms of action (MOA), optimizing compound efficacy, and mitigating off-target effects. Drug Affinity Responsive Target Stability (DARTS) is a powerful, label-free technique that enables precise characterization of protein–ligand interactions under physiologically relevant conditions.
Creative Proteomics provides a comprehensive, customizable DARTS assay service tailored to pharmaceutical, biotechnology, and academic researchers seeking to validate drug targets or investigate compound selectivity and engagement profiles. Our platform is engineered to deliver high-resolution, reproducible, and quantifiable data, helping streamline target discovery workflows.
Why Choose DARTS Analysis?
Addressing Key Client Needs: Mechanism Validation | Target Deconvolution | Drug Efficacy Assessment
DARTS leverages the principle that ligand binding stabilizes the conformation of target proteins, making them more resistant to proteolysis. By comparing the proteolytic degradation profiles of compound-treated versus untreated samples, researchers can infer direct binding events—even in complex mixtures such as whole cell lysates.
This technique is particularly valuable when:
- Elucidating Mechanisms of Action (MOA) for phenotypic screening hits
- Deconvoluting Targets of bioactive compounds with unknown binding partners
- Profiling Target Engagement under native conditions
- Comparing Affinities of analog molecules across a target panel
- Identifying Off-Target Effects in safety and toxicology assessments
DARTS is non-biased, label-free, and applicable across a broad range of small molecules and protein classes, including kinases, transcription factors, and epigenetic regulators.
Comprehensive DARTS Assay Services Offered by Creative Proteomics Creative Proteomics
Creative Proteomics provides an end-to-end DARTS analysis solution, combining rigorous sample preparation, proteolytic digestion optimization, and mass spectrometry-based protein identification. Our modular service suite includes:
- Target Engagement Analysis: Quantitative assessment of compound–protein binding and stabilization profiles
- Comparative Proteolysis Profiling: High-throughput analysis of drug-induced stability shifts across treatment conditions
- Binding Affinity Ranking: Dose-dependent DARTS profiling to rank compound–target affinities
- Custom Protease Selection: Multiple protease options (e.g., Pronase, Thermolysin) tailored to target properties and compound characteristics
- Native Lysate Screening: Target analysis within intact, untreated lysates for true physiologic relevance
- LC-MS/MS Identification & Quantification: High-confidence detection of stabilized proteins using advanced proteomics
- Data Analysis & Bioinformatics Support: Statistical enrichment analysis, GO term annotation, and pathway visualization
All services are customizable based on client objectives, species, compound solubility, or target expression systems (e.g., mammalian cells, bacterial lysates, or tissue homogenates).
Advantages of Our DARTS Assay Platform?
- Label-Free Design: Avoids genetic modification, fusion tags, or chemical labels that may alter native protein conformations
- High Sensitivity: Capable of detecting binding-induced stability shifts for proteins in the low nanomolar to micromolar range
- Physiological Relevance: Assay conducted under near-native buffer and temperature conditions
- Parallel Screening: Ability to assess hundreds of potential targets simultaneously
- Quantitative Correlation: Stabilization degree correlates with binding affinity, allowing compound ranking
- Broad Compatibility: Suitable for diverse small molecule classes including kinase inhibitors, GPCR modulators, and PROTACs
Proven Tools for High-Confidence Target Identification
To ensure precise and reproducible DARTS profiling, Creative Proteomics integrates robust instrumentation platforms:
Proteolytic Digestion
Controlled proteolysis using Pronase, Thermolysin, or Subtilisin based on target stability and protease compatibility
Separation and Detection
- Peptide separation using Thermo Scientific Vanquish™ UHPLC system
- Detection with Orbitrap Fusion Lumos Tribrid Mass Spectrometer for ultra-high resolution (up to 500,000 FWHM)
- Optional: Bruker timsTOF Pro for enhanced ion mobility separation and increased depth of coverage
Data Analysis Tools
- Protein identification via MaxQuant and Proteome Discoverer
- Quantitative differential analysis for downstream interpretation
- Customized reports including volcano plots, target enrichment scores, and mechanistic pathway mapping

Vanquish™ UHPLC system (Fig from Thermo Scientific)

Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Fig from Thermo Scientific)
Workflow for DARTS Assay
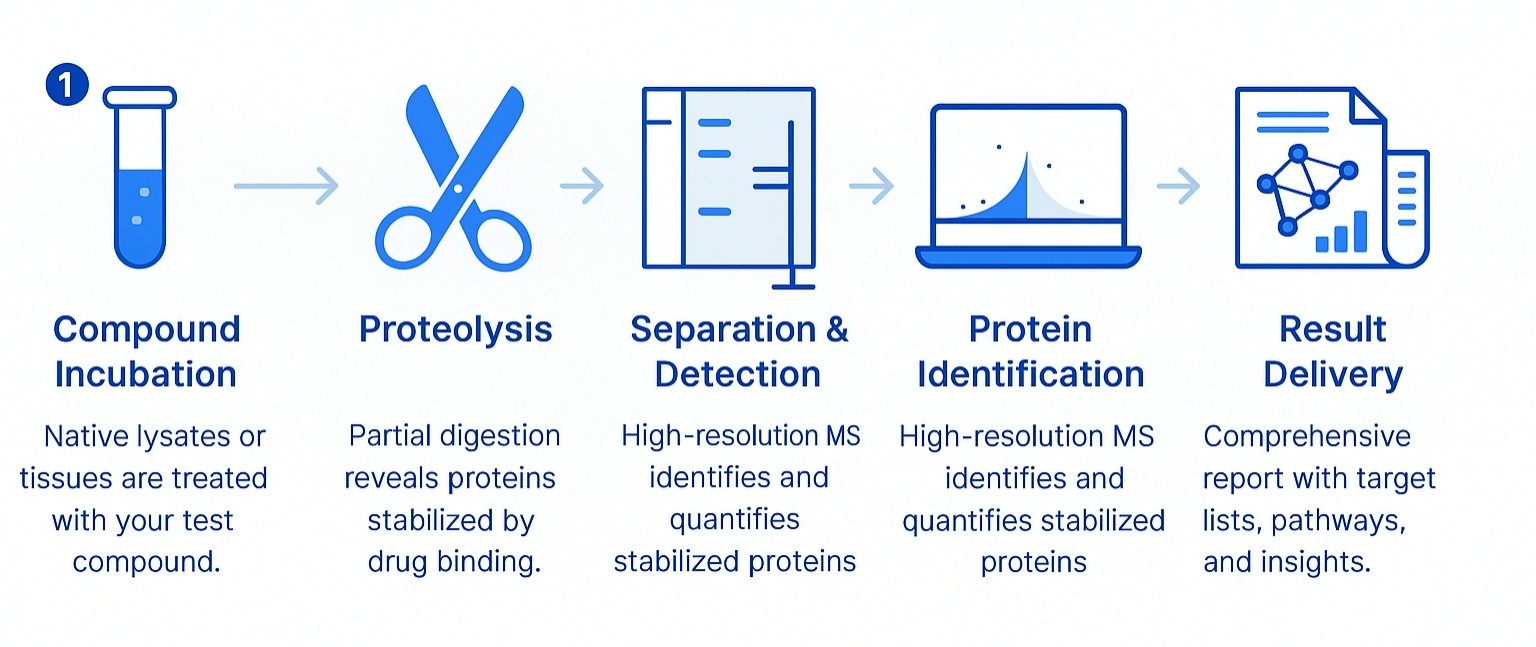
Sample Requirements for DARTS Analysis Service
Below is a summary of acceptable sample types and minimum input quantities for successful DARTS assay execution:
| Sample Type | Minimum Sample Volume | Notes |
|---|---|---|
| Whole Cell Lysate (Mammalian) | ≥ 500 µg total protein | Preferred for most small molecule profiling; ensure detergent-free buffer |
| Tissue Homogenate | ≥ 1 mg total protein | Snap-frozen or freshly prepared; homogenization buffer compatibility required |
| Bacterial Lysate | ≥ 500 µg total protein | Suitable for target discovery in microbial systems |
| Purified Protein (optional) | ≥ 50 µg per condition | For orthogonal validation of interaction specificity |
| Drug Compound | ≥ 100 µL of 10 mM DMSO stock | Provide solubility, MW, and any stability information |
Note: All samples should be stored at –80°C and shipped on dry ice. Buffer exchange may be necessary if incompatible detergents or salts are present.
Demo Results– Validating Drug–Target Interactions with Confidence
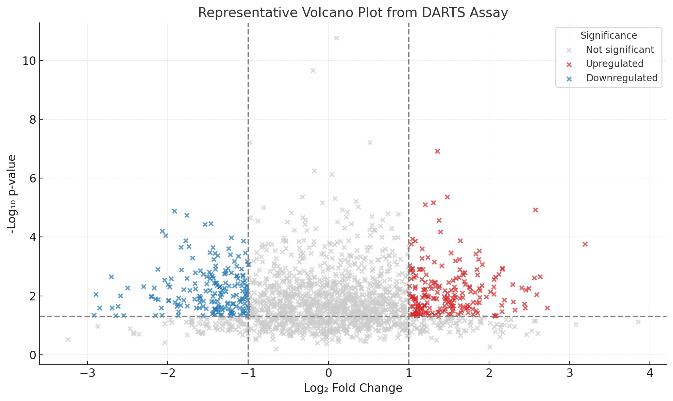
Global Stabilization Profile
A volcano plot reveals clear differentiation between stabilized and unaffected proteins after compound treatment.
- 1,800+ proteins quantified
- 86 significantly stabilized targets (log₂FC ≥ 1.0, p < 0.05)
- Enrichment in pathways related to cell cycle, apoptosis, and proteostasis
Example Target Hits
In a small-molecule kinase project, DARTS identified:
- HSP90: log₂FC = 2.7, p = 0.0008
- CDK2: log₂FC = 1.9, p = 0.0021
These hits aligned with expected biological activity and informed downstream validation.
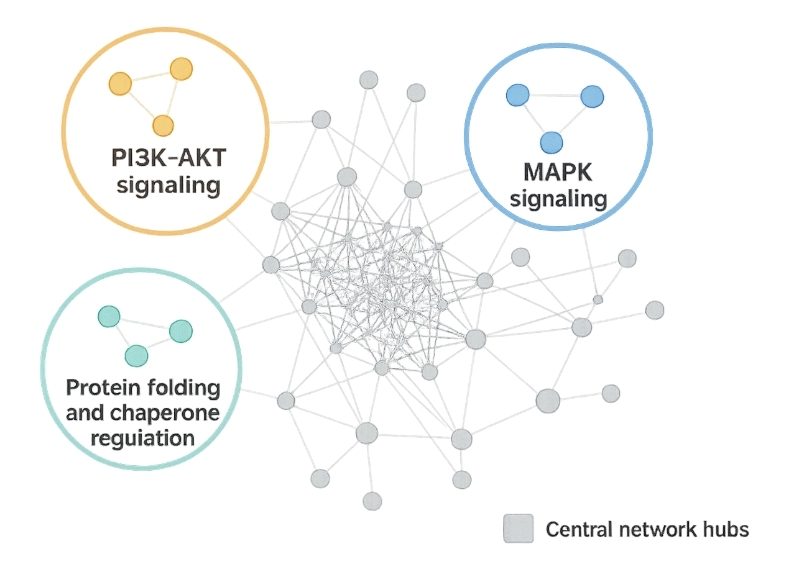
Functional Pathway Analysis
GO and pathway enrichment highlighted:
- PI3K-AKT and MAPK signaling
- Protein folding and chaperone regulation
- Central network hubs via STRING PPI mapping
Platform Performance Summary
| Metric | Result |
|---|---|
| Quantified proteins | 1,500–2,300 |
| Avg. sequence coverage | 65–85% |
| Significant stabilized hits | 50–150 per compound |
| Instruments used | Orbitrap Fusion Lumos / timsTOF Pro |
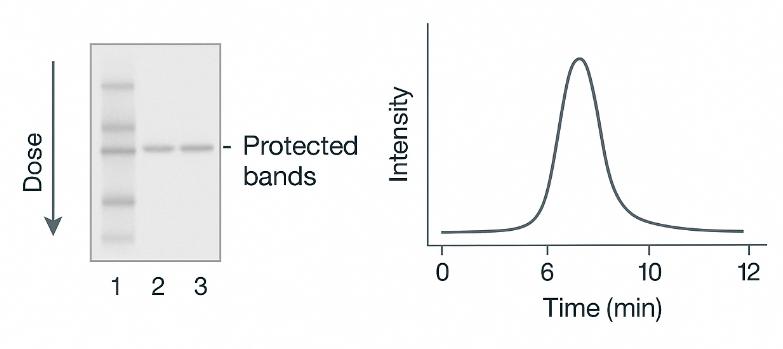
Experimental Validation
SDS-PAGE and LC-MS profiles confirm compound-induced protection of target proteins across doses, ensuring biological relevance before large-scale quantitation.
FAQ of DARTS Assay Service
How does DARTS differentiate between direct binders and downstream stability effects caused by signaling or metabolic changes?
DARTS is conducted in lysates or cell extracts, typically in the absence of ATP or other cofactors that drive post-translational modifications. This eliminates indirect, pathway-mediated effects, enabling identification of direct physical interactions based on proteolytic resistance. However, to further confirm specificity, we recommend complementing DARTS with dose–response curves and competition assays using inactive analogs.
How is protease type and digestion time optimized for different targets or compounds?
Protease selection is critical. We offer a panel including Pronase, Thermolysin, and Subtilisin, each with different cleavage site preferences and structural tolerance. For example:
- Pronase: broad-spectrum; ideal for global screening
- Thermolysin: heat-stable; suited for thermophilic targets
- Subtilisin: mild cleavage; better for partially folded or membrane proteins
Digestion kinetics are optimized for each matrix to achieve partial proteolysis, preserving differential stability patterns without over-digestion. Pilot experiments are conducted as needed.
How are protein abundance and stabilization effects quantitatively assessed in DARTS?
We use label-free quantification (LFQ) or TMT/iTRAQ labeling (upon request) via high-resolution MS. Stabilization is evaluated by comparing peptide intensity or spectral counts between treated and control samples. Fold changes are statistically evaluated using tools such as limma or MSstats, and visualized via volcano plots or heatmaps. A Z-score or p-value threshold is used to define significantly stabilized proteins.
How do you address non-specific stabilization or false positives due to aggregation or off-target binding?
We mitigate false positives by:
- Including vehicle controls and compound analogs for specificity screening
- Using replicates to assess reproducibility
- Filtering out known aggregators or sticky proteins (e.g., via CRAPome database)
- Applying bioinformatic filtering to cross-reference DARTS hits with known target families, molecular weight, or expected binding domains
Can DARTS be used to analyze allosteric or covalent interactions?
Yes. DARTS is capable of detecting both orthosteric and allosteric interactions, as long as the binding event alters the structural stability of the protein. For covalent binders, stabilization is often more pronounced, but detection depends on protease accessibility and binding site location. Additional mapping strategies (e.g., limited proteolysis followed by targeted MS/MS) can be used for detailed analysis.
What is the sequence coverage typically achieved for target proteins, and how does that affect interpretation?
With LC-MS/MS on Orbitrap Fusion Lumos or timsTOF Pro, we typically achieve 50–85% sequence coverage for abundant and moderately expressed proteins. For low-abundance proteins, we recommend fractionation or targeted MS methods (e.g., PRM). Coverage depth directly affects the confidence in stabilization calls, especially for small shifts or isoform-specific effects.
Can Creative Proteomics support DARTS studies for biologics, such as peptides, PROTACs, or antibody fragments?
Yes. We routinely work with non-small molecule binders, including:
- Peptides and macrocycles
- Targeted degraders (e.g., PROTACs)
- Bi-functional molecules
For PROTACs, DARTS can indirectly capture ternary complex formation by identifying E3 ligase co-enrichment or unexpected stabilization of target and partner proteins. Complementary assays such as co-IP or other orthogonal validation methods can then be arranged to confirm these findings.
What bioinformatics tools are used to interpret functional relevance of stabilized targets?
We integrate DARTS results into functional pathway contexts using:
- GO enrichment (Gene Ontology Biological Process, Molecular Function)
- Reactome and KEGG pathway mapping
- STRING for protein–protein interaction networks
- Custom scoring matrices to correlate DARTS hits with compound class or phenotypic outcomes
How does DARTS compare with other target ID methods like TPP, or chemoproteomics?
| Method | Binding Type | Environment | Quantitative | Strengths |
|---|---|---|---|---|
| DARTS | Direct binding (stabilization) | Lysates | Yes | Label-free, native conditions, broad proteome coverage |
| TPP | Thermal profiling + MS | Cells or lysates | Yes | Multiplexed thermal shift curves |
| ABPP | Covalent binding only | Cells | Yes | Probe-based, selective for nucleophilic residues |
DARTS complements these methods and is especially powerful when structural stabilization is the main measurable outcome of binding, even in complex matrices.
Learn about other Q&A about proteomics technology.




