β-Amino Acids Analysis Service
Get isomer-aware, defensible quantification of β-amino acids in complex matrices—separating α/β positional isomers and D/L enantiomers—with study-ready QC and clear deliverables from Creative Proteomics.
- Isomer-specific LC–MS/MS: HILIC/mixed-mode; optional chiral LC or Marfey's
- Sensitivity: compound-dependent LOD 0.5–5 ng/mL, LOQ 2–15 ng/mL
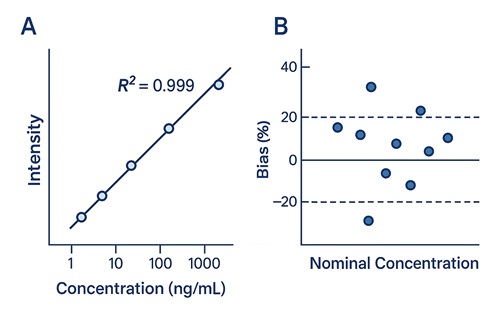
- Quant performance: R² ≥ 0.995, accuracy/precision within ±15% (±20% at LOQ)
- Acquisition rigor: scheduled MRM/PRM; ≥2 transitions + qualifier-ratio checks
- Throughput: plate-compatible prep; 6–12 min per sample methods
- Matrix coverage: plasma, CSF, urine, tissue, cells, fermentation broth
Submit Your Request Now
×
What You Will Receive
- Raw data package: vendor files + mzML
- Quant tables: concentrations/response ratios with RT & qualifier ratios (CSV/XLSX)
- Calibration & QC summary: R², bias/CV, LOQ/weighting, acceptance checks
- Method sheet: LC gradients, columns, MRM/PRM transitions, collision energies
- Isomer/Chiral report: α/β resolution evidence; optional D/L ratios
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQs
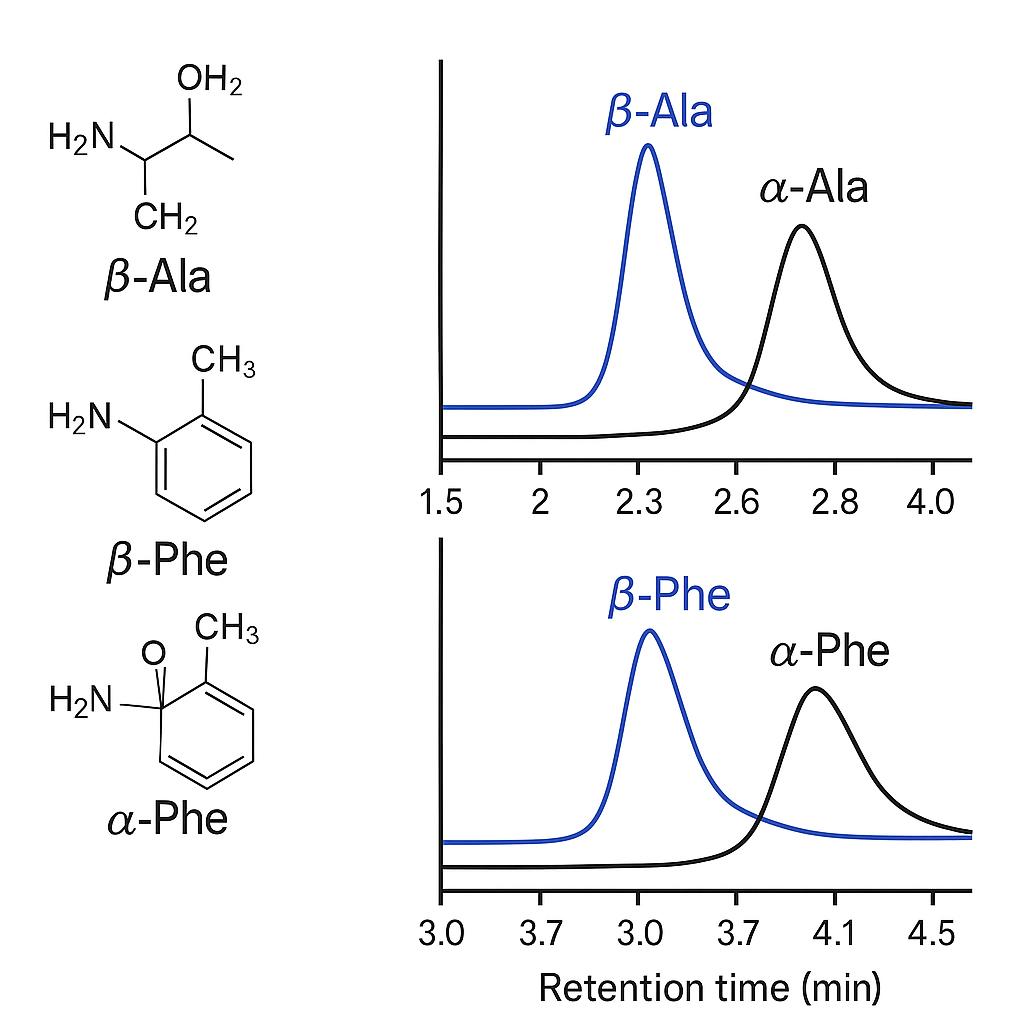
What Are β-Amino Acids and Why Analyze Them?
β-Amino acids are amino acids in which the amino group is attached to the β-carbon (one carbon further from the carboxyl group than in α-amino acids). This structural shift alters acid–base properties, transport, and catabolic routes. Many β-AAs are chiral, exist as L/D enantiomers, or as constitutional isomers relative to α-AAs (e.g., β-Phe vs α-Phe). Accurate measurement therefore requires isomer-aware chromatography, appropriate derivatization (when used), and MS-level selectivity to avoid coelution with abundant α-AAs or matrix amines.
What Problems We Solve for Our Clients
- Isomer Interference: α/β isobars and coeluting amines cause false positives. We implement orthogonal separation (HILIC or mixed-mode; optional chiral columns) with MRM confirmation to remove ambiguity.
- Poor Sensitivity in Complex Matrices: Zwitterions can tail and ionize weakly. We stabilize and enrich analytes, then apply tuned source conditions and isotope-labeled internal standards to reach low-ng/mL detection ranges (compound-dependent).
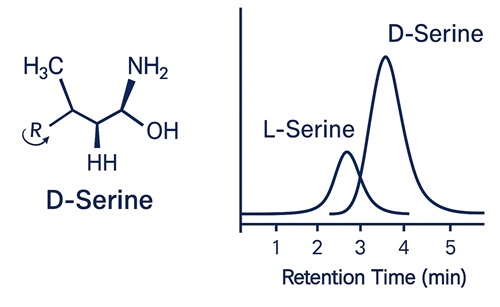
- Chiral Questions: When biological interpretation requires enantiomeric ratios, we provide direct chiral LC–MS/MS or Marfey's-type derivatization for D/L resolution.
- Reproducibility Across Batches: We run bracketed calibrators, matrix-matched QCs, and pooled QCs with acceptance criteria (typ. ≤15% CV intra-batch; ≤20% inter-batch) to secure trendable data.
β-Amino Acids Analysis Services Offered by Creative Proteomics
- Targeted Biothiols Panel (quantitative): Cys, Cys-Gly, Hcy, γ-Glu-Cys, GSH/GSSG, CoA-SH, NAC, Cystine, Homocystine
- Extended Sulfur Metabolites (semi-quant/quant): Cysteamine, 3-mercaptopropionic acid, 2-mercaptoethanol, penicillamine
- Volatile β-Amino Acids Fingerprinting (targeted GC–MS/MS): 2-furfurylthiol, 4MMP, 3MH, 3MHA, MFT, benzyl mercaptan, ethanethiol
- Protein Thiol/Disulfide State (mapping): Differential alkylation and peptide-level LC–MS for relative thiol occupancy
- Redox Metrics: GSH/GSSG, Cys/Cystine, Hcy/Homocystine ratios and Nernst-based redox potential (optional)
- Stability & Stress Studies: Thiol loss kinetics under process conditions; antioxidant/additive screening
β-Amino Acids Analysis Targets & Compound Panels
| Class | Analyte | Chiral? | Ion Mode | Typical LOQ(biofluid)* | Preferred Separation |
|---|---|---|---|---|---|
| Aliphatic β-Amino Acids | β-Alanine (3-Aminopropionic acid) | No | ESI+ | 2–10 ng/mL | HILIC / derivatized RP |
| BAIBA (β-Aminoisobutyric acid; AIBA) | Yes (D/L) | ESI+ | 5–15 ng/mL | Chiral LC / Marfey's | |
| BABA (β-Aminobutyric acid; 3-ABA) | Yes | ESI+ | 5–15 ng/mL | HILIC / Mixed-mode | |
| β-Aminovaleric acid (3-Aminopentanoic acid) | Yes | ESI+ | 5–15 ng/mL | HILIC | |
| Dicarboxylic β-Amino Acids | β-Aspartic acid (3-Aminosuccinic acid) | Yes | ESI− / + | 10–30 ng/mL | Mixed-mode / HILIC |
| β-Glutamic acid (3-Aminoglutaric acid) | Yes | ESI− / + | 10–30 ng/mL | Mixed-mode / HILIC | |
| Aromatic β-Amino Acids | β-Phenylalanine | Yes | ESI+ | 5–20 ng/mL | Mixed-mode |
| β-Tyrosine | Yes | ESI+ | 5–20 ng/mL | Mixed-mode | |
| β-Tryptophan (3-Amino-3-indolepropionic acid) | Yes | ESI+ | 10–30 ng/mL | Mixed-mode / RP | |
| Branched β-Amino Acids | β-Leucine | Yes | ESI+ | 10–25 ng/mL | Chiral LC |
| β-Isoleucine | Yes | ESI+ | 10–25 ng/mL | Chiral LC | |
| Sulfur-Containing β-Amino Acids | β-Methionine | Yes | ESI+ | 10–30 ng/mL | RP / Mixed-mode |
| β-Cysteine (β-Mercaptoalanine) | Yes | ESI+ | 10–30 ng/mL | HILIC | |
| Hydroxy β-Amino Acids | β-Serine | Yes | ESI+ | 10–25 ng/mL | HILIC |
Advantages of Our β-Amino Acids Analysis Services
- Sensitivity: Compound-dependent LOD typically 0.5–5 ng/mL in biofluids with isotope-dilution; LOQ typically 2–15 ng/mL (validated per matrix/analyte).
- Linearity: R² ≥ 0.995 across multi-point calibration (≥6 levels) with back-calculated bias ≤15% (≤20% at LOQ).
- Precision & Accuracy: Intra-batch CV ≤ 10–12%; inter-batch CV ≤ 15–20% with matrix-matched QCs.
- Recovery & Matrix Effect: Spike-recovery 85–115%; post-extraction matrix factor monitored and corrected via stable-isotope IS (e.g., β-Ala-d3, BAIBA-d3).
- Carryover & Stability: Carryover<0.1% of high standard; processed-sample stability verified up to typical batch runtime; freeze–thaw tolerance profiled (on request).
- Throughput: 96-well compatible prep; run times as short as 6–12 min per sample (panel-dependent) to support medium-to-large studies.
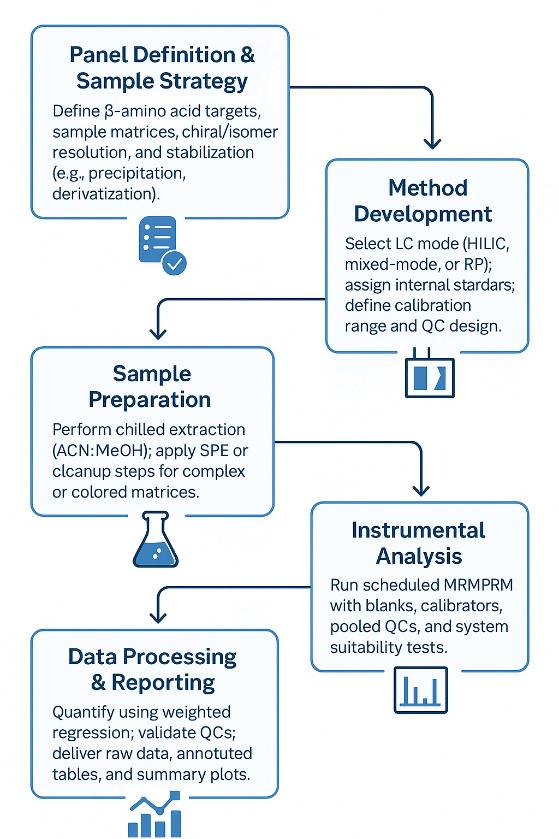
Workflow for β-Amino Acids Analysis Service
1. Scoping & Panel Design — define compounds, matrices, expected ranges, chiral/isomer needs, and reporting format.
2. Stabilization Strategy — choose protein precipitation vs. on-receipt derivatization; define pH and temperature controls to suppress transamination/degradation.
3. Method Setup — select HILIC/mixed-mode vs. derivatized RP; assign isotope-labeled IS; set calibration range and QC levels.
4. Extraction & Cleanup — chilled precipitation (e.g., ACN:MeOH); optional SPE for pigment/salt removal (environmental/fermentation).
5. Acquisition — scheduled MRM or PRM with system suitability checks, blanks, bracketed QCs, and pooled matrix controls.
6. Quantification & QA — regression (1/x or 1/x² weighting); QC acceptance vs. predefined criteria; batch review for drift and carryover.
7. Reporting & Handover — raw files, mzML, transition list, quantified tables (with IS, RT, qualifiers), QC summary plots, and an interpretation-focused memo.
Technology Platform for β-Amino Acids Analysis Service
Creative Proteomics primarily uses LC–MS/MS–based targeted quantification workflows, with method setups tailored to β-amino acids' polarity, isomer complexity, and matrix background. The most commonly used configurations include:
Instrument Platforms
- UHPLC systems: Thermo Vanquish, Waters ACQUITY I-Class
- MS detectors: SCIEX QTRAP 6500+, Agilent 6495C
- Ion source: Electrospray Ionization (ESI), typically in positive mode
Chromatographic Modes
- HILIC: Preferred for native β-amino acids due to their high polarity
- Mixed-Mode LC: Enables α/β isomer discrimination and matrix decomplexing
- Chiral LC (optional): Used when enantiomer (D/L) separation is required
Derivatization (if needed)
- Reagents like AQC or FMOC-Cl are used for hydrophobic enhancement and reversed-phase compatibility
- Marfey's reagent (FDAA) is applied when D/L enantiomer resolution via RP-LC is preferred
Acquisition & Quantification
- Scheduled MRM (Multiple Reaction Monitoring) with ≥2 transitions per analyte
- LOQs typically range from 2–15 ng/mL depending on compound and matrix
- Quantification uses weighted (1/x or 1/x²) regression with matrix-matched calibrators and QCs
- Qualifier/quantifier ion ratios and retention time match ensure compound identity

SCIEX Triple Quad™ 6500+

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Waters ACQUITY UPLC System
Sample Requirements for β-Amino Acids Analysis Service
| Matrix | Minimum Amount | Pretreatment / Notes | Storage & Shipping | Rejection Criteria |
|---|---|---|---|---|
| Plasma/Serum | 80–120 µL | Prefer K2-EDTA; avoid hemolysis; no heparin (ion suppression) | Freeze at −80 °C; ship on dry ice | Thawed on arrival; hemolysis; unknown anticoagulant |
| Urine | 200 µL | Record creatinine if normalization needed | −80 °C; dry ice | Preservatives added without disclosure |
| CSF | 60–100 µL | Handle cold; minimize freeze–thaw | −80 °C; dry ice | Visible particulates/contamination |
| Cell Pellet | ≥2×10^6 cells or ≥10 mg wet | Wash PBS (ice-cold), snap-freeze | −80 °C; dry ice | Buffer salts/detergents >0.1% |
| Tissue | ≥20 mg | Snap-freeze; annotate region | −80 °C; dry ice | Formalin-fixed or thawed |
| Fermentation Broth / Media | 1–2 mL | Filter or clarify if viscous | −80 °C; dry ice | Unknown additives/biocides |
Demo Results
FAQ of β-Amino Acids Analysis Service
What is the minimum quantifiable level of β-amino acids in complex matrices?
Our LC–MS/MS methods typically reach low-ng/mL quantification levels, depending on compound structure and matrix complexity. Sensitivity is enhanced using isotope-labeled standards and matrix-matched calibration.
Can your platform distinguish β-amino acids from their α-isomeric or structural analogs?
Yes. We apply mixed-mode or orthogonal HILIC methods with retention time confirmation and qualifier ion ratios to resolve β-/α-isomers. Method specificity is analytically verified against known interferents.
How do you ensure the accuracy and reproducibility of β-amino acid quantification across batches?
Each run includes multi-level matrix QCs, bracketed calibrators, and pooled sample controls. Inter-batch CV is typically maintained within acceptable bioanalytical thresholds using a predefined acceptance plan.
Is D-/L-enantiomeric resolution available for chiral β-amino acids?
Yes. Enantiomer-specific analysis is available through direct chiral LC–MS or Marfey's derivatization. Enantiomeric ratios can be reported upon request with full resolution validation.
How do you handle matrices with low β-amino acid abundance or strong interference?
We offer targeted enrichment protocols, matrix-specific SPE cleanup, and background subtraction strategies. Additionally, interference-prone matrices can be processed using modified LC gradients and internal standard compensation.
What types of matrices are supported for β-amino acid profiling?
Typical matrices include plasma, CSF, urine, cell lysates, microbial broth, fermentation supernatants, and tissue homogenates. Extraction and calibration strategies are adapted per matrix type.
Do you support high-throughput β-amino acid screening in multi-sample projects?
Yes. We offer 96-well compatible workflows with short-cycle methods optimized for batch-scale acquisition while preserving quantitation quality. QC design is scalable across large studies.
Can I customize the panel to include rare or pathway-specific β-amino acids?
Custom panels are supported. You may submit a compound list or pathway interest area, and we will assess method feasibility, expected detection limits, and required reference standards.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the metabolomics-related papers published by our clients:

- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023.
- The activity of the aryl hydrocarbon receptor in T cells tunes the gut microenvironment to sustain autoimmunity and neuroinflammation. 2023.
- Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. 2023.
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023.
- Anti-inflammatory activity of black soldier fly oil associated with modulation of TLR signaling: A metabolomic approach. 2023.
- Non-invasive elevation of circulating corticosterone increases the rejection of foreign eggs in female American robins (Turdus migratorius). 2022.
- Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking. 2023.
- Nicotine exposure during rodent pregnancy alters the composition of maternal gut microbiota and abundance of maternal and amniotic short chain fatty acids. 2022.
- Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium. 2023.