Title: Glucosylceramide is essential for Heartland and Dabie bandavirus glycoprotein-induced membrane fusion
Journal: PLOS Pathogens
Published: 2023
Background
Emerging tick-borne zoonotic viruses, including Heartland bandavirus (HRTV) and Dabie bandavirus (DBV), have become a public health concern due to their potential to cause hemorrhagic fever and high fatality. Despite the serious health risks, the mechanisms underlying HRTV and DBV infections are not well understood. Both viruses rely on their surface glycoproteins, Gn and Gc, for entry into host cells. Recent studies have highlighted the role of glycosphingolipids, such as glucosylceramide (GlcCer), in the viral lifecycle. This study identified the glycosphingolipid biosynthesis pathway as essential for HRTV and DBV infection, revealing that GlcCer is required for viral entry. The research also demonstrated that GlcCer interacts with the Gc protein to facilitate membrane fusion, providing insight into potential therapeutic targets for these viral infections.
Materials & Methods
Cells, Viruses, and Reagents
- Cell lines: HEK293T, HeLa, and Sf9 insect cells were cultured in DMEM (with 10% FBS) and ExpiSf CD medium for insect cells, maintained at 37°C in a 5% CO₂ incubator.
- Viruses: Heartland virus (HRTV) MO4 strain and recombinant vesicular stomatitis virus (rVSV) strains were used for infection studies.
- Antibodies: Flag antibody, β-actin, LAMP1, and antibodies against HRTV N-protein were purchased from various vendors for western blot and immunofluorescence assays.
- Reagents: NB-DNJ (glucosylceramide synthase inhibitor), UGT8 inhibitor 19, various lipids (e.g., GlcCer, GalCer, DOPC) were obtained for lipidomics analysis and lipid interaction studies.
Lipidomics Analysis (SFC-MS/MS)
- Lipid Extraction: Lipids were extracted from cells using a modified Bligh and Dyer method, with chloroform/methanol/water (2:1:0.8) as the solvent system. Cell pellets from infected and control cells were homogenized in this solvent mixture. After phase separation, the organic phase containing lipids was collected and evaporated under nitrogen.
- Lipid Identification: Lipid species were identified and quantified using Supercritical Fluid Chromatography-Tandem Mass Spectrometry (SFC-MS/MS). Lipid extracts were reconstituted in methanol and analyzed by direct infusion into the mass spectrometer. Lipids of interest, such as glucosylceramide (GlcCer) and galactosylceramide (GalCer), were detected based on their specific molecular ions and fragmentation patterns. The lipid content was quantified using internal standards, and lipid profiles were compared between virus-infected and uninfected cells.
- Data Analysis: Lipidomic data were processed and analyzed using lipidomic analysis software to identify significant changes in lipid composition in response to HRTV infection. Relative lipid abundance was calculated, and heatmaps were generated to visualize lipid alterations. Pathway analysis was performed to investigate potential lipid pathways involved in viral entry and replication.
CRISPR-Cas9 Knockout Screening for Lipid Pathway Genes
- Library: The GeCKO v2A genome-wide CRISPR library was utilized to knockout genes related to lipid metabolism, including those involved in ceramide biosynthesis and glycosphingolipid production.
- Screening Process: HEK293T cells were transduced with the CRISPR library, followed by selection with puromycin and subsequent infection with HRTV. After a recovery period, genomic DNA was isolated, and sgRNA sequences were amplified and subjected to next-generation sequencing.
- Analysis: Gene enrichment analysis was performed using MAGeCK software to identify genes that significantly affected the HRTV replication cycle. Pathway enrichment analysis was performed using DAVID to identify lipid-related pathways that contribute to HRTV infection.
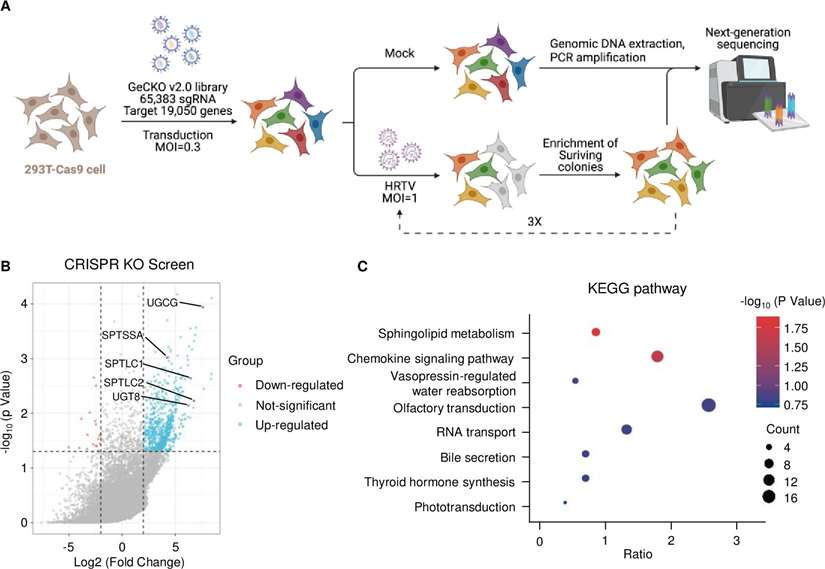 Genome-wide CRISPR screen identifies HRTV entry factors.
Genome-wide CRISPR screen identifies HRTV entry factors.
Immunofluorescence and Confocal Microscopy
- Virus Localization: For immunofluorescence studies, cells were infected with HRTV, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Cells were incubated with primary antibodies against HRTV N protein and visualized with secondary antibodies conjugated to Alexa Fluor dyes.
- Lipid Staining: Lipid-rich regions were labeled using fluorescent lipid analogs (e.g., BODIPY FL C5-ceramide) to visualize lipid raft domains. Confocal microscopy was used to assess the co-localization of viral proteins with specific lipids at the cell membrane or within cellular compartments.
Liposome Preparation and Protein-Lipid Binding Assay
- Liposome Generation: Liposomes containing different lipid compositions (e.g., DOPC, GlcCer, GalCer) were prepared by film hydration followed by extrusion through a polycarbonate membrane to obtain liposomes of uniform size (~100 nm).
- Protein-Lipid Interaction: Recombinant HRTV Gc protein was expressed in insect cells and purified. Liposome binding assays were performed by incubating purified Gc protein with lipid vesicles. Protein binding to the liposomes was quantified using a fluorescent lipid displacement assay. The binding affinity between Gc protein and lipids was determined by surface plasmon resonance (SPR) or biolayer interferometry (BLI).
Subcellular Fractionation
- Fractionation: To study lipid-protein interactions at the subcellular level, cells were fractionated into plasma membrane, endosomal, and cytosolic fractions by differential centrifugation. Lipid content in each fraction was determined by lipidomic analysis (SFC-MS/MS), and the localization of HRTV Gc protein was assessed by western blotting.
- Lipid Interaction: To study the interaction of lipids with viral proteins, lipid binding assays were performed on isolated membrane fractions. The presence of ceramide species and other lipids was assessed in relation to HRTV protein localization.
Data Analysis and Statistical Methods
- Lipidomic Data Analysis: Quantification of lipid species was performed using internal standards, and statistical significance between experimental groups was determined by Student's t-test or ANOVA using GraphPad Prism software. Lipid abundance was compared between infected and uninfected cells, and pathway enrichment was analyzed to identify lipid pathways modulated by viral infection.
Results
Glucosylceramide Biosynthesis Pathway is Essential for HRTV Infection:
- A genome-wide CRISPR/Cas9 knockout screen in 293T-Cas9 cells identified 755 genes enriched after HRTV infection. The sphingolipid metabolism pathway, particularly the enzymes involved in GlcCer biosynthesis (UGCG, UGT8), was critical for HRTV replication.
- Inhibiting GlcCer synthesis (with NB-DNJ) reduced HRTV replication, while UGT8 inhibition had no such effect.
- GlcCer depletion in cells resulted in reduced viral replication and resistance to infection, confirming the importance of the sphingolipid pathway.
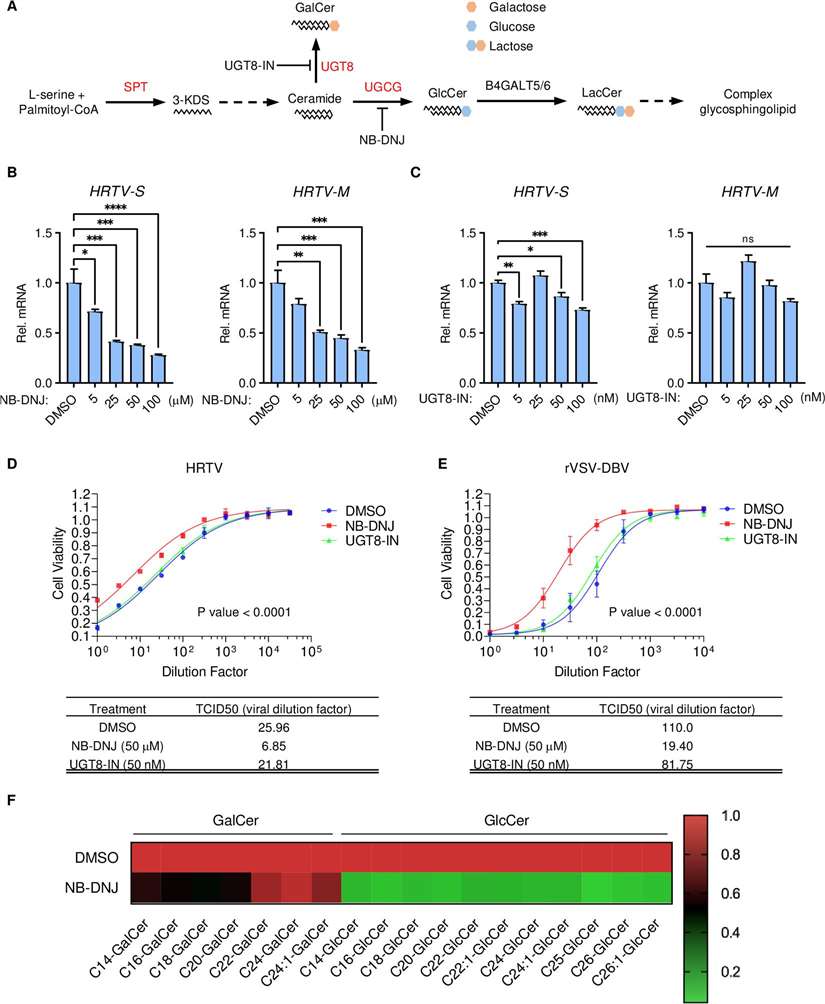 Glucosylceramide biosynthesis pathway is essential for HRTV infection.
Glucosylceramide biosynthesis pathway is essential for HRTV infection.
Role of GlcCer in HRTV Infection:
- CRISPR/Cas9 knockout of sphingolipid synthesis genes (SPTLC2, UGCG, UGT8) showed varying degrees of inhibition in HRTV infection.
- GlcCer levels were directly correlated with infection efficiency, as cells deficient in GlcCer (e.g., UGCG-KO) exhibited reduced infection, whereas B4GALT5/6 knockout cells, with higher GlcCer levels, showed enhanced infection.
- Supplementing GlcCer in knockout cells rescued HRTV infection, establishing the critical role of GlcCer in viral entry.
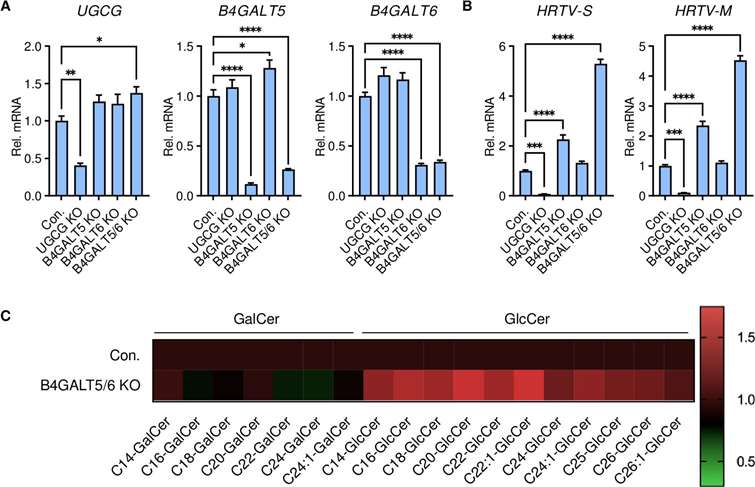 Glycosphingolipid downstream of GlcCer is not required for HRTV infection.
Glycosphingolipid downstream of GlcCer is not required for HRTV infection.
GlcCer and Membrane Fusion:
GlcCer deficiency did not affect the binding or internalization of HRTV, but it disrupted the trafficking of the virus from endosomes to the cytosol.Membrane fusion assays showed that GlcCer was essential for efficient HRTV-induced membrane fusion, as GlcCer-deficient cells exhibited reduced fusion activity.
HRTV or DBV Glycoproteins and GlcCer-Mediated Membrane Fusion:
HRTV and DBV glycoproteins (Gn and Gc) were found to mediate entry through GlcCer. The entry of recombinant viruses pseudotyped with HRTV or DBV glycoproteins was significantly reduced in GlcCer-deficient cells, which could be rescued by GlcCer supplementation.
Interaction between Gc Protein and GlcCer:
HRTV Gc protein directly interacts with GlcCer in the lipid bilayer, facilitating membrane fusion.
Surface plasmon resonance and bio-layer interferometry assays showed stronger binding of HRTV Gc to GlcCer compared to GalCer or phosphatidylcholine, highlighting the critical role of GlcCer in viral entry.
The Role of D841 Residue in Gc Protein for GlcCer-Mediated Entry:
Mutation of the D841 residue in the Gc protein's lipid-binding pocket disrupted its ability to bind GlcCer, leading to a significant reduction in viral infectivity.
GlcCer supplementation could not rescue the infectivity of mutants, confirming that the D841 residue is essential for the Gc protein's lipid interaction and viral entry.
Reference
- Xia, Tian, et al. "Glucosylceramide is essential for Heartland and Dabie bandavirus glycoprotein-induced membrane fusion." PLoS Pathogens 19.3 (2023): e1011232.




