Oxalic Acid Analysis Service – Precise Quantification for Metabolomics Research
At Creative Proteomics, our oxalic acid analysis service delivers precise quantification of oxalic acid and its related metabolites using advanced HPLC and LC-MS/MS platforms. We help researchers overcome the challenge of measuring low-abundance organic acids, ensuring accurate, reproducible results across food science, plant metabolism, environmental chemistry, and metabolic pathway analysis.
- Solve the problem of inconsistent oxalic acid measurements
- Ensure reproducibility with isotope-labelled internal standards
- Support targeted metabolomics research with publication-ready outputs
Submit Your Request Now
×
- Why It Matters
- Applications
- Methods
- Pathway Analysis
- Workflow
- Advantages
- Sample Requirements
- FAQ
- Case Study
- Publications
Why Oxalic Acid Analysis Matters
Oxalic acid is a central organic acid that provides insights into both biochemical and environmental processes. In plants, it influences calcium binding, carbon metabolism, and stress responses. In microbial systems and soils, oxalic acid plays a role in nutrient cycling and mineral interactions. For researchers, precise oxalic acid determination is a key step in understanding metabolic regulation and pathway activity.
Accurate analysis supports:
- Food and nutrition studies evaluating oxalate content in plant-derived foods.
- Metabolomics research connecting oxalic acid metabolism to glyoxylate and TCA pathways.
- Environmental studies investigating microbial metabolism and soil chemistry.
- Biotechnology optimising organic acid production in fermentation systems.
Applications of Oxalic Acid Profiling
Food and Nutrition Research
Oxalic acid content affects nutrient absorption and food quality. Profiling oxalate levels in leafy vegetables, cereals, or fermented foods enables researchers to evaluate food composition and trace how processing methods alter bioavailability. Metabolomics methods have become integral to advancing molecular nutrition and food analysis.
Plant Metabolism and Biotechnological Improvement
Oxalic acid plays a dual role in plants: it can act as a defence metabolite or impair food quality depending on context. Studies highlight its importance in stress response, mineral regulation, and as a target for genetic or processing interventions to reduce anti nutrient effects.
Soil and Environmental Research
In soils and microbial ecosystems, oxalic acid influences nutrient cycling and mineral solubilisation. Profiling oxalate contributes to understanding soil chemistry and microbial metabolism, with implications for agroecology and bioprocessing.
Industrial and Sustainable Oxalic Acid Production
Beyond metabolite profiling, oxalic acid is also valuable as a bioproduct in various industries—ranging from textiles to metal recovery. Biotechnological approaches using microbial fermentation emphasize sustainability and environmental benefits
Oxalic Acid Analysis Methods Comparison
| Method | Strengths & Features | Typical Applications |
|---|---|---|
| HPLC (UV/RID, Ion-Exclusion) | Reliable separation using mixed-mode or ion-exclusion columns. Strong UV or RI detection; suitable for complex samples. | Food matrices, plant tissue, fermentation broths |
| LC-MS/MS | High sensitivity and specificity. Derivatization-based kits enable harmonized quantitation across labs. | Environmental samples, metabolomics, soil and hydroponic extracts |
| Spectrophotometric / Colorimetric | Low-cost, rapid assays. Example: diazotization methods for oxalate detection. | High-throughput screening and basic lab analyses |
| Titrimetric / Gravimetric | Classical, equipment-free quantification; precipitation and redox approaches. | Educational purposes, preliminary estimates |
| Emerging Techniques (GC-MS, Electrochemical, CE) | Specialized options: volatile compound profiling via GC-MS; electrochemical or electrophoretic methods for unique samples. | Niche research needs, advanced laboratories |
Oxalic Acid Metabolic Pathway Analysis
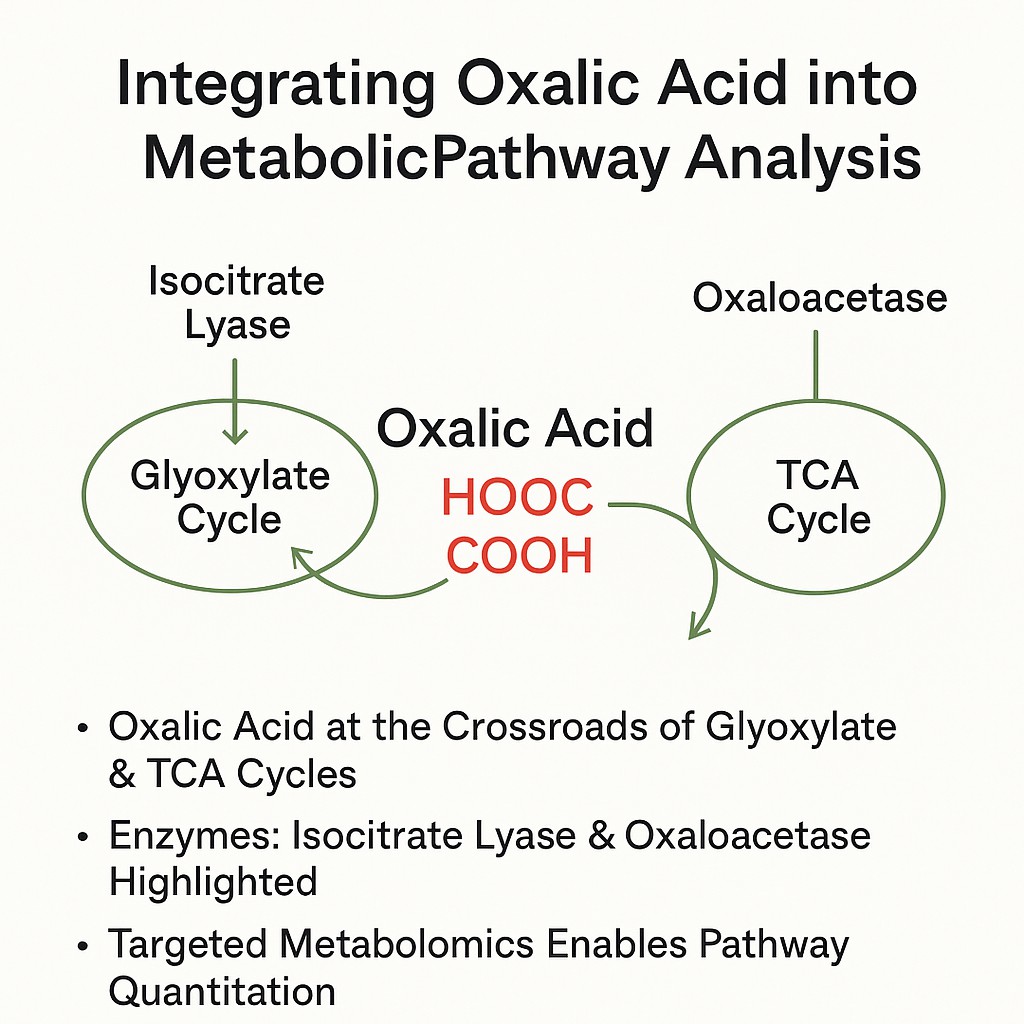
- Positions oxalic acid at the intersection of the glyoxylate cycle and TCA pathway, showing its role in central carbon metabolism.
- Core enzymes such as isocitrate lyase, oxaloacetase (OXA), and glyoxylate dehydrogenase (GLOXDH) mediate biosynthesis and degradation of oxalic acid.
- In fungi, oxalate derives from precursors in the TCA or glyoxylate pathways via cytoplasmic or peroxisomal enzyme activity.
- Secreted oxalate influences soil mineral transformation and nutrient cycling, adding ecological value to pathway analysis.
Targeted metabolomics enables quantitative mapping of the oxalic acid metabolic pathway, enhancing insight into pathway regulation and flux.
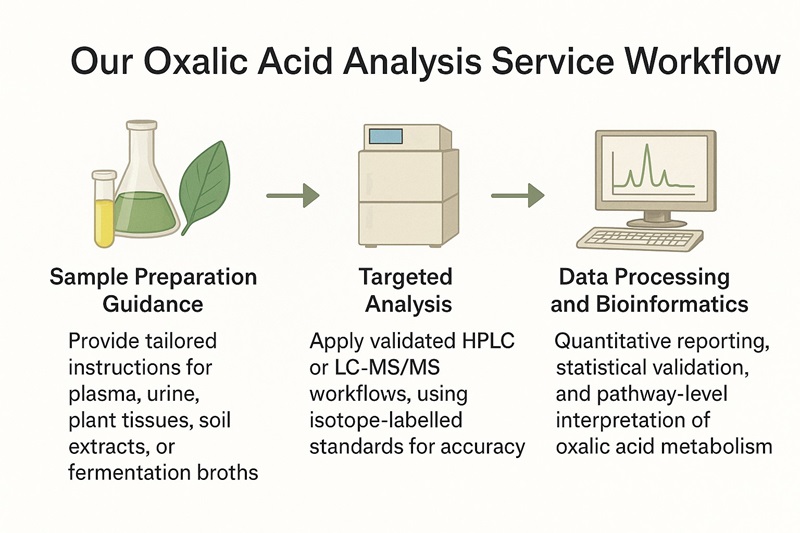
Our Oxalic Acid Analysis Service Workflow
Project Consultation
Define analytical scope, detection methods, and pathway integration goals with our experts.
Sample Preparation Guidance
Provide tailored instructions for plasma, urine, plant tissues, soil extracts, or fermentation broths.
Targeted Analysis
Apply validated HPLC or LC-MS/MS workflows, using isotope-labelled standards for accuracy.
Data Processing and Bioinformatics
Quantitative reporting, statistical validation, and pathway-level interpretation of oxalic acid metabolism.
Publication-Ready Reporting
Deliverables include chromatograms, quantitative tables, and expert commentary aligned with journal requirements.
Advantages of Choosing Creative Proteomics
Partnering with Creative Proteomics ensures that every oxalic acid analysis project benefits from advanced technology, expert guidance, and reproducible results.
Validated Analytical Platforms
HPLC and LC-MS/MS platforms deliver high sensitivity and specificity for oxalic acid determination.
Isotope-Labelled Standards
Guarantee accurate quantification even at trace levels across diverse sample types.
Comprehensive Coverage
Capable of measuring free oxalate, bound oxalate, and pathway-related metabolites.
Tailored Solutions
Flexible workflows adapted to plant, food, environmental, and targeted metabolomics studies.
Sample Requirements for Oxalic Acid Analysis
| Sample Type | Recommended Amount | Notes / Pre-treatment |
|---|---|---|
| Animal Tissue | 100–200 mg | Remove blood residues with pre-chilled water; snap-freeze in liquid nitrogen; store at –80 °C. |
| Plant Tissue | 100–200 mg | Collect specific sections; wash quickly if needed; snap-freeze in liquid nitrogen; store at –80 °C. |
| Plasma / Serum | >100 µL | Collect, centrifuge, snap-freeze in liquid nitrogen; store at –80 °C. |
| Urine | 200–500 µL | Centrifuge at 3000 rpm for 10 min at 4 °C; collect supernatant; freeze in liquid nitrogen; store at –80 °C. |
| Saliva / Bile / Amniotic fluid / Tears | >200 µL | Centrifuge to remove debris; freeze in liquid nitrogen; store at –80 °C. |
| Cells | >1 × 10⁷ | Wash with pre-cooled PBS; collect cell pellet (≥50 µL); snap-freeze; store at –80 °C. |
| Culture Supernatant | >2 mL | Centrifuge to remove cells/debris; freeze in liquid nitrogen; store at –80 °C. |
| Microbial Culture | >2 mL (liquid) or pellet | Collect by centrifugation; wash with pre-cooled PBS; snap-freeze pellet; store at –80 °C. |
| Feces / Intestinal Content | 100–200 mg | Aliquot immediately after collection; freeze in liquid nitrogen; store at –80 °C. |
| Soil Sample | >1 g | Freeze-dry, sieve through 2 mm; store at –80 °C. |
| Swab Samples | 2 swabs | Collect with sterile swabs, store at –80 °C; avoid contamination. |
Key Submission Notes
- All samples should be flash-frozen in liquid nitrogen immediately after collection and stored at –80 °C.
- Avoid repeated freeze–thaw cycles.
- Use screw-cap cryovials for transport; ship on dry ice.
- Biological replicates recommended:
- Microorganisms & Plants: ≥ 6–8 replicates
- Model organisms: ≥ 8–10 replicates
- Clinical samples: ≥ 30 replicates
Frequently Asked Questions (FAQ)
What is the most effective method for oxalic acid analysis?
The most effective methods are HPLC and LC-MS/MS, with LC-MS/MS offering higher sensitivity and precision, especially when using isotope-labelled standards for low-level detection—a preferred approach for targeted metabolomics panels.
Which sample types can be analysed for oxalic acid?
Validated sample types include plasma, urine, plant tissues, soil extracts, fermentation broths, microbial cultures, and related matrices—each prepared according to protocol to ensure stability and compatibility with chromatography and mass spectrometry workflows.
Can oxalic acid analysis measure both free and bound forms?
Yes, our workflows quantify both free oxalic acid and bound oxalates following proper sample extraction and preparation to capture the complete oxalate profile in the selected sample type.
Do you include data interpretation and pathway context in the results?
Yes, every oxalic acid analysis report includes quantitative results, quality control data, and context through metabolic pathway interpretation—ideal for integrating into broader pathway analysis or targeted metabolomics.
Can oxalic acid profiling be combined with other metabolomics services?
Absolutely, oxalic acid analysis can be bundled with broader organic acid profiling or targeted metabolomics services to enable holistic metabolite mapping and support metabolic pathway analysis.
Learn about other Q&A about proteomics technology.
Client Case Study

Comparative Metabolite Profiling of Salt Sensitive Oryza sativa and Halophytic Wild Rice Oryza coarctata Under Salt Stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I. Plant‐Environment Interactions. 2024.
https://doi.org/10.1002/pei3.10155
- Background
- How We Helped
- Results
- Impact
Salinity is a critical abiotic stress that limits rice productivity worldwide. The halophytic wild rice Oryza coarctata shows remarkable salt tolerance compared to the cultivated, salt-sensitive Oryza sativa. To explore the biochemical basis of this adaptation, the authors performed root-specific untargeted metabolomics using LC–MS, supported by Creative Proteomics workflows. Organic acids, including oxalic acid and related metabolites, were a focus due to their roles in osmotic adjustment and stress regulation.
Using our LC–MS untargeted metabolomics platform, we provided high-resolution metabolite profiling across control and salt-stress conditions. Over 1000 metabolites, including organic acids, fatty acyls, amino acids, and sugars, were identified. Bioinformatics analysis integrated PCA, clustering, and KEGG pathway enrichment to highlight differences in metabolic strategies between the two rice species.
- O. coarctata maintained higher basal levels of key metabolites (e.g., vanillic acid, threonic acid, amino acids) under control conditions, forming a metabolic "buffer" against salt stress.
- In contrast, O. sativa showed strong upregulation of organic acids only after salt exposure, reflecting a reactive rather than pre-emptive strategy.
- Pathway enrichment highlighted differences in glyoxylate and dicarboxylate metabolism, phenylpropanoid biosynthesis, and organic acid metabolism, pathways closely linked with oxalic acid dynamics.
- The PCA plot on page 3 shows clear separation of metabolite profiles between control and salt stress conditions in both species.
- The volcano plots and bar charts on pages 4–5 illustrate significantly modulated metabolites, including differential accumulation of organic acids and stress-related compounds (Figure 2–4).
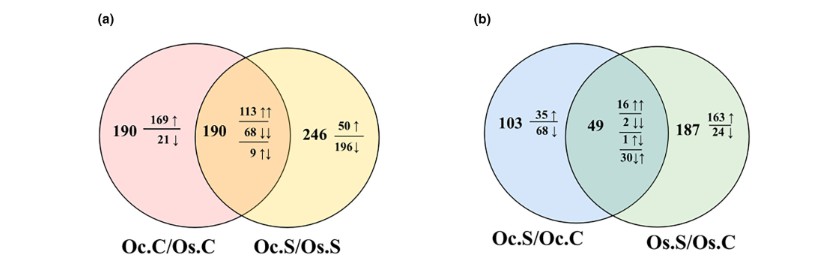 FIGURE 2 Venn diagrams of differentially accumulated metabolites
FIGURE 2 Venn diagrams of differentially accumulated metabolites
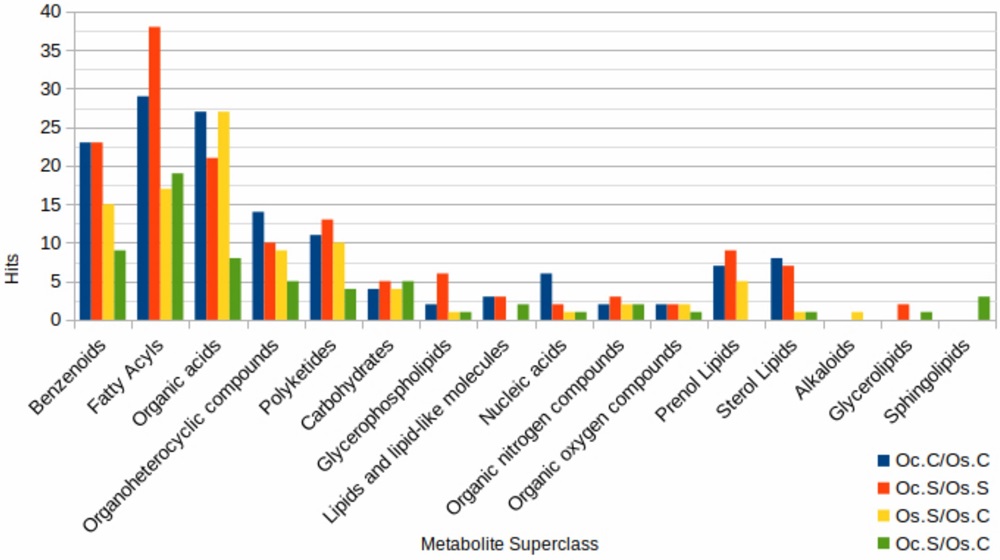 FIGURE 3 Grouped bar plot indicating enriched metabolite sets analysis in four comparison groups
FIGURE 3 Grouped bar plot indicating enriched metabolite sets analysis in four comparison groups
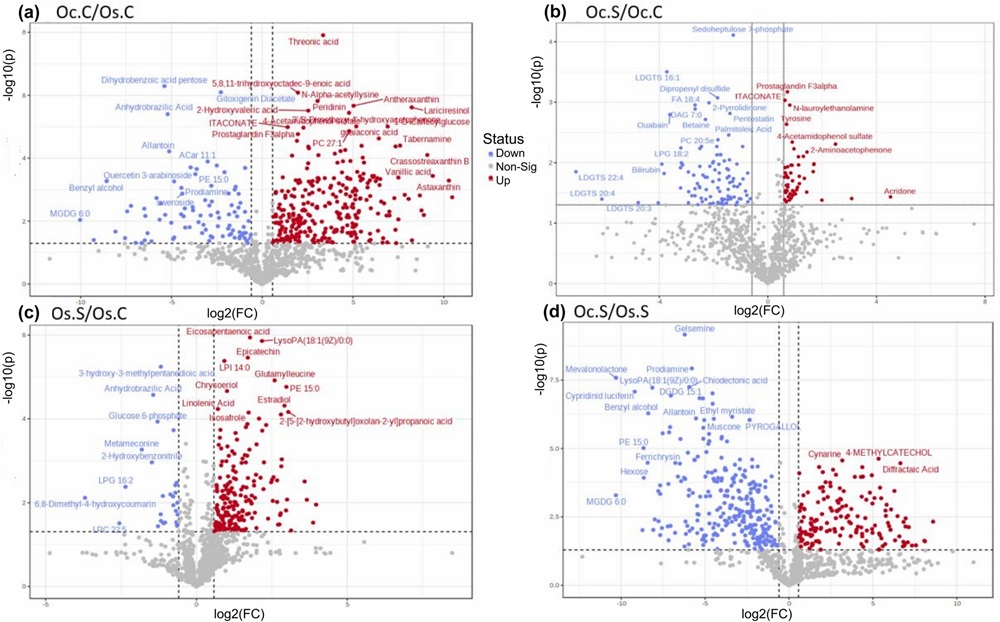 FIGURE 4 Volcano plots representing significantly modulated metabolites in comparison groups
FIGURE 4 Volcano plots representing significantly modulated metabolites in comparison groups
This case study demonstrates the power of oxalic acid analysis and broader organic acid profiling in identifying stress tolerance mechanisms. By revealing contrasting metabolic responses, the study provides targets for metabolic engineering and crop breeding programs aimed at enhancing salt resistance in rice. It exemplifies how Creative Proteomics' metabolomics services support pathway-level discovery and translational research.
Publications

- High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes. Transplantation and Cellular Therapy. 2024.
- Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content. Proceedings of the National Academy of Sciences. 2024.
- The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner. International Journal of Molecular Sciences. 2024.














