Top-Down Protein Sequencing
- Home
- Services
- Protein Sequencing
- Mass Spectrometry Based Protein Sequencing
- Top-Down Protein Sequencing
Service Details
Top-down protein sequencing is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable identifying and quantitating unique proteoforms through the analysis of intact proteins. During mass spectrometry, electrospray ionization typically are used to ionize the intact proteins that then are trapped in a Fourier transform ion cyclotron (Penning trap), quadrupole ion trap (Paul trap) or Orbitrap mass spectrometer. Usually, fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Top-down MS proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.
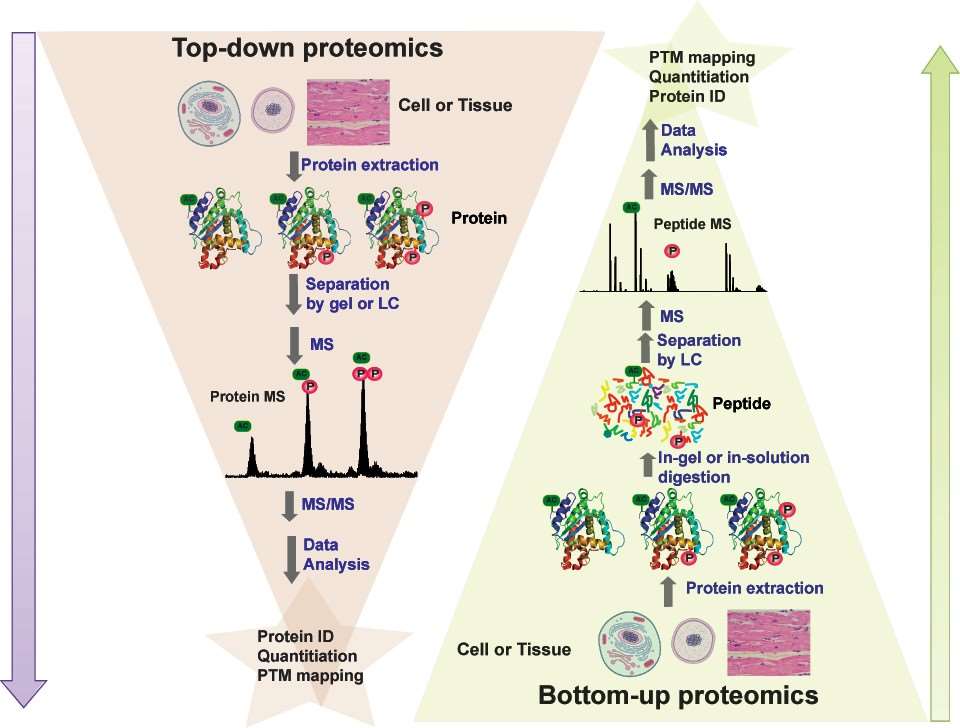 Fig.
1. Schematic illustration of the difference between protein-based top-down and peptide-based bottom-up proteomics.
(Gregorich Z R, et al., 2014)
Fig.
1. Schematic illustration of the difference between protein-based top-down and peptide-based bottom-up proteomics.
(Gregorich Z R, et al., 2014)
Creative Proteomics provides protein sequencing analysis services based on the Top down method, and MALDI ISD mass spectrometry technology was used to provide N-terminal and C-terminal sequencing services. Compared to the traditional Edman degradation method, Top-down protein sequencing can be performed even when the end of protein are modified, including N-terminal acetylation, N-terminal pyroglutamate, sequence truncation and signal sequence, or blocked.
Top-down protein sequencing pipeline we provided is that intact proteins are first separated from complex biological samples by reversed-phase liquid chromatography and then directly ionized by matrix-assisted laser desorption/ionization (MALDI) techniques. The resulting ions are fragmented by collision-induced dissociation (CID), high-energy collision-induced dissociation (HCD), electron capture dissociation (ECD), or electron transfer dissociation (ETD) and analyzed in tandem mass spectrometry.
Please feel free to contact us If you would like to discuss the best way to determine N/C sequencing of your protein. At Creative Proteomics, we not only provide professional and comprehensive top-down protein sequencing service projects for global scientific researchers, our scientists can also provide and help to design the best solutions according to your specific requirements, and describe them in detailed project reports, we look forward to with your cooperation.
References
For research use only, not intended for any clinical use.