IgA Antibody Sequencing
- Home
- Services
- Antibody Sequencing
- Mass Spectrometry Based Antibody Sequencing
- Antibody Full Amino Acid Sequencing
- IgA Antibody Sequencing
Service Details
Immunoglobulin A (IgA) plays a key role in defending mucosal surfaces against attack by infectious microorganisms. Indeed, IgA is not just the most prevalent antibody class at mucosal sites, but is also present at signifificant concentrations in serum. The unique structural features of the IgA heavy chain allow IgA to polymerise, resulting in mainly dimeric forms, along with some higher polymers, in secretions In humans, there are two subclasses of IgA, named IgA1 and IgA2. Like all Ig, each subclass comprises a basic molecular unit of two identical heavy chains (HCs) and two identical light chains (LCs). Each chain begins at its N-terminus with a variable region, which is followed by a constant region. The LCs are the same in each subclass, but the HCs diffffer within their constant regions, which are encoded by distinct Cα genes. Two allotypic variants of human IgA2, known as IgA2m(1) and IgA2m(2), have been characterised. A third IgA2 variant, termed IgA2(n), has been described, but while presumed to be an allelic form, its penetrance in the population remains to be investigated.
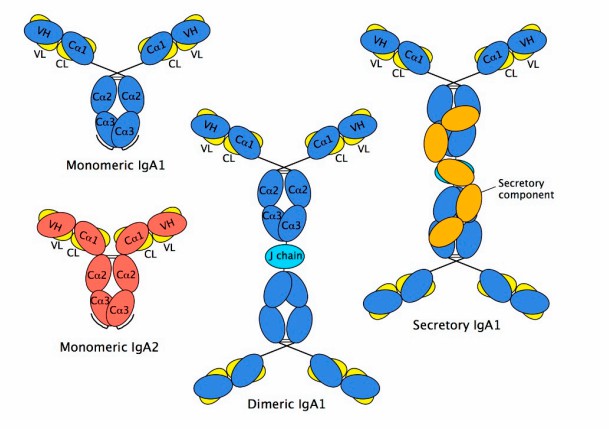 Fig.
1. Schematic diagram of IgA structures-monomeric, dimeric, and secretory IgA. (Nejadmoghaddam M R, et al.,
2019)
Fig.
1. Schematic diagram of IgA structures-monomeric, dimeric, and secretory IgA. (Nejadmoghaddam M R, et al.,
2019)
Creative Proteomics has developed an advanced immunoglobulin profiling sequencing method that uses HCD + ETD to generate simple, easy-to-read sequence ladders for IgA light and heavy chains. Developed by our scientists with extensive antibody sequencing experience Compared with the traditional sequencing method, the antibody de novo sequencing method of the present invention adopts the ionization method of ETD combined with HCD, which increases the formation of fragment ions, which is suitable for deformed and natural intact antibodies.
At Creative Proteomics, we use HCD + ETD fragmentation to increase coverage. In addition, our scientists can use the "Database Assisted Shotgun Sequencing" (DASS) technology to combine the advantages of existing technologies to perform excellent, fast and precise sequencing services for immunoglobulin antibodies. When similar proteins and known genome sequences exist, we are able to integrate de novo peptide, intensity and positional confidence scores from databases and homology to find protein sequences with high accuracy.
We have sequenced dozens of immunoglobulin A (IgA) antibodies. Customers only need to provide 50-100 µg of IgA sample to obtain the amino acid sequence information of the antibody and ensure 100% sequence accuracy.
Creative Proteomics provides professional IgA antibody sequencing services for global customers. Our team has successfully sequenced thousands of antibodies to help researchers decode their antibodies in a fraction of the time. Therefore, please contact us if you are interested in our antibody sequencing services.
References
For research use only, not intended for any clinical use.