Single Molecule Fluorescence Sequencing
- Home
- Services
- Protein Sequencing
- Single Molecule Fluorescence Sequencing
Service Details
Single-molecule fluorescence sequencing is a method of DNA sequencing. With the development of this technology, a new approach to protein sequencing has been derived from fluorescence-based single-molecule protein sequencing. The principle is to first decompose the protein into peptide fragments, selectively label one or more specific amino acids in the peptide fragments with specific fluorescent groups, and observe the fluorescence on different amino acids, so as to determine the partial amino acid sequence of the peptide fragments, and then the These fluorescent amino acid sequences were compared to a reference proteome to determine their protein origin. Single-molecule protein sequencing is considered to be a promising new method in the field of proteomics. Like nanopore sequencing, it can detect very small protein samples. It is a combination of DNA sequencing and mass spectrometry proteomics. Novel protein sequencing technology with classical Edman sequencing.
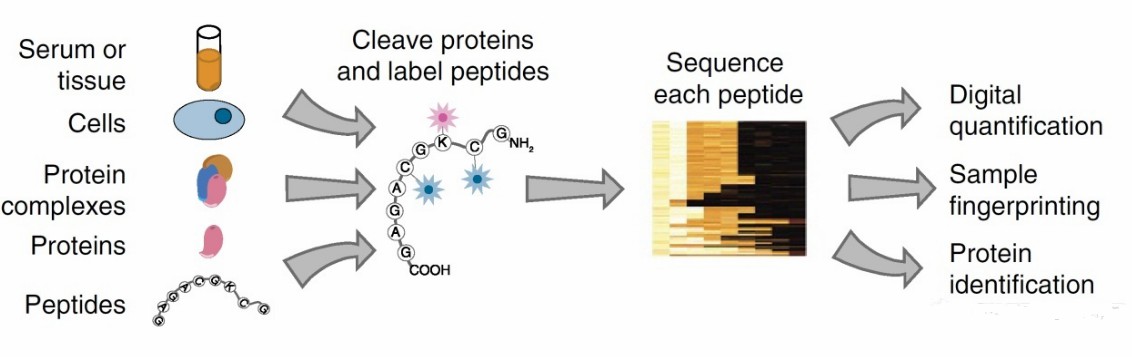 Fig. 1. Overview of
single-molecule fluorosequencing. (Swaminathan J, et al., 2018)
Fig. 1. Overview of
single-molecule fluorosequencing. (Swaminathan J, et al., 2018)
As a rapidly growing global protein sequencing service provider, Creative Proteomics has a team of highly qualified scientists focused on developing emerging fluorescence-based protein molecular sequencing solutions. At Creative Proteomics, our professional scientists have developed a single molecule fluorescent protein sequencing technology that tags several amino acids, including lysine and cysteine, with specific fluorophores. We implement fluorescent sequencing of single-molecule proteins by labeling and discriminating simple peptide mixtures with peptides.
Here, we provide a set of workfolw of single-molecule fluorosequencing:
Ⅰ. One or more enzymes are used to enzymatically digest target protein to form a large number of peptide fragments.
Ⅱ. A fluorescent group, usually an N-terminal specific amino acid binding agent, is selected to selectively label one or a specific amino acid in the peptide segment.
Ⅲ. Multiple rounds of Edman degradation are performed sequentially to remove the N-terminal amino acid, monitoring the changing fluorescence intensity to generate a fluorescent signature for unique identification of individual peptides.
Ⅳ. Different amino acid sequences marked with fluorescence are observed to determine the partial amino acid sequence of the peptide by image processing algorithm.
Ⅴ. The measured amino acid sequence and the amino acid sequence group are compared to obtain the target protein amino acid sequence.
With our protein sequencing experience, at Creative Proteomics, you also get:
Our highly qualified scientists develop a complete sequencing pipeline for you to decode your protein. At Creative Proteomics, we can offer you far more than single molecule fluorescence sequencing. Using our protein sequencing service can be the start of a long. But if you have any special requirement, you are welcome to contact us for custom service. We look forward to your happy cooperation.
References
For research use only, not intended for any clinical use.