Protein Sumoylation Identification Service
- Home
- Applications
- Proteomics Analysis Services
- Protein Post-Translational Modification Analysis Services
- Protein Sumoylation Identification Service
Service Details
Small ubiquitin-like modifier (SUMO) is a post-translational modification of protein, which refers to the covalent attachment of a small ubiquitin-related modifier (SUMO) to the protein. A protein with approximately 18% homology to ubiquitin, the SUMO protein is approximately 100 amino acids in length and has considerable structural overlap with ubiquitin. In mammals, the SUMO family includes four known SUMO isomers: SUMO 1-4.
The conjugation of SUMO, known as SUMOylation, is an essential PTM that is highly conserved in eukaryotes and is involved in regulating the subcellular localization of many substrate proteins, such as gene transcription, subnuclear structure formation, viral infection, and cell cycle progression. The imbalance of SUMOylation and deSUMOylation is related to the occurrence and progression of various diseases, and the identification of protein SUMO will help to elucidate disease pathogenesis and provide support to develop potential therapeutic strategies or target drugs.
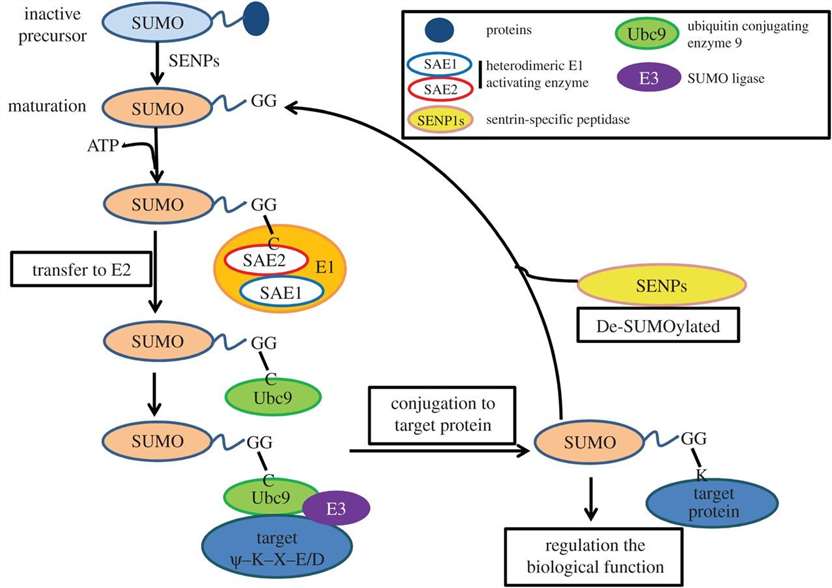 Fig. 1.
Biochemical process of SUMO modifications in mammal cells. (Yang Y, et al., 2017)
Fig. 1.
Biochemical process of SUMO modifications in mammal cells. (Yang Y, et al., 2017)
The methods of SUMOylation identification mainly include bioinformatics combined with amino acid site-directed mutagenesis and mass spectrometry (MS)-based proteomics analysis. With a powerful mass spectrometry sequencing platform, Creative Proteomics has launched a high-sensitivity mass spectrometry analysis platform for protein SUMOylation identification. Multiple samples and sumoylation in eukaryotes and prokaryotes can be analyzed for robust identification and quantification of SUMO modifications.
At Creative Proteomics, our highly qualified scientists isolate target SUMOylated proteins by affinity chromatography, and then identify them by MS. Throughout the service cycle, SUMO proteins were digested with endoprotease Lys-C, enriched using specific antibodies or affinity-immobilized metal affinity chromatography, and identified by MS.
With cutting-edge equipment and expertise in protein sequencing and mass spectrometry, we provide one-stop protein sumoylation identification service. You only need to tell us the purpose of your experiment and send your samples to us, we will take care of all the follow-up matters of the project.
The simple process of our service includes:
Thanks to our powerful mass spectrometry sequencing platform, Creative Proteomics provides a one-stop protein sumoylation identification service for customers worldwhile. Our experienced scientists work with you to develop tailor-made analytical solutions. Please feel free to contact us with any questions regarding protein sumoylation identification.
References
For research use only, not intended for any clinical use.