Protein Structure Identification Service
- Home
- Applications
- Proteomics Analysis Services
- Protein Identification Services
- Protein Structure Identification Service
Service Details
Protein structure is the result of non-covalent interactions of amino acid residues in three-dimensional space. Typically, proteins fold into one or more specific spatial conformations to perform their biological functions. Protein structure involves four different levels, namely primary structure, secondary structure, tertiary structure, and quaternary structure. Among them, primary structure is the basis of protein biological function, and its determination and structure are the premise of understanding protein structure and function. The determination of protein primary structure mainly includes the determination of the number of polypeptide chains, the type, number and arrangement of amino acids in each polypeptide chain, as well as the position and number of intra- or inter-chain disulfide bonds in polypeptide chains. Nowdays, with the development of analytical tools, mass spectrometry has become a powerful tool for protein determination. In proteomic studies, the technology can even be used to sequence protein mixtures. However, in most cases, a combination of techniques is used to produce the most reliable results.
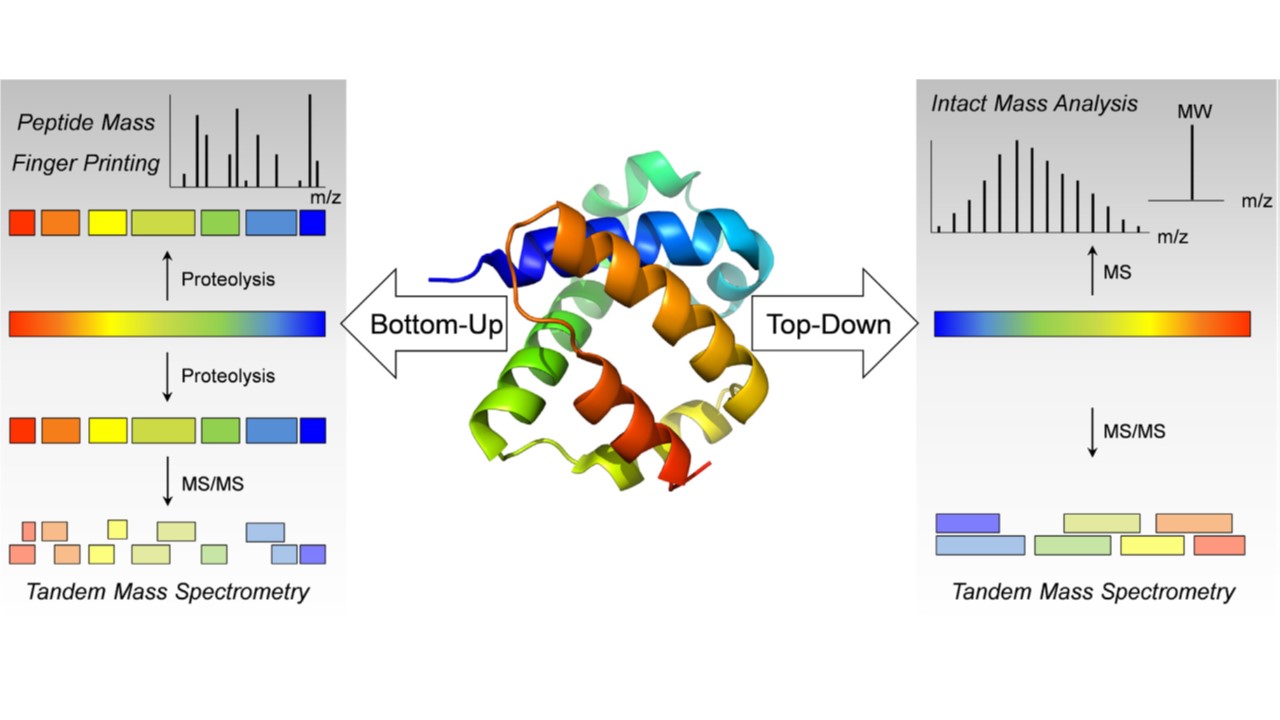 Fig.
1. Bottom-up and top-down analysis for the structural characterization of peptides and protein pharmaceuticals by
mass spectrometry. (Leurs U, et al., 2015)
Fig.
1. Bottom-up and top-down analysis for the structural characterization of peptides and protein pharmaceuticals by
mass spectrometry. (Leurs U, et al., 2015)
Structural analysis of a protein usually begins with the determination of the amino acid sequence of its primary structure, and the determination of primary structure is the first most necessary and critical step you take in protein research for drug development.
At Creative Proteomics, with many years of sequencing experience, our protein structure identification services are able to meet customer requirements for specific targets, and the successful implementation of this service relies on peptide mapping MS/MS analysis and de novo sequencing technology developed by my scientists.
During protein sequence analysis, six proteases (trypsin, chymotrypsin, Asp-N, Gluc-C, Lys-C, and Lys-N) are used by our highly skilled to digest your protein of interest and the resulting peptides can be reliably isolated and identified segment, then drill down to the complete sequence information of the protein. During this process, while peptide fragments are obtained by mass spectrometry, 100% certainty of the protein sequence is done by splicing between peptides. Each amino acid composition and surrounding amino acid microenvironment, including disulfide bond information, can then be displayed.
Here, our experienced scientists provide you with comprehensive and reliable protein primary structure analysis techniques, some of the outstanding services include:
In accordance with the regulatory requirements of ICH Q6B, we use a combination of molecular mass spectrometry (MS) and peptide mapping analysis to determine your protein's primary structure (amino acid sequence).
Additionally, our experienced professional scientists will use de novo sequencing techniques to characterize your protein's primary structure (amino acid sequence), when the protein sequence was not available in open protein or genome databases.
Thanks to our powerful mass spectrometry sequencing platform, Creative Proteomics, as a first-class protein sequencing service provider, is committed to providing global customers with all protein structure identification service to accelerate their project research journey. Our services guarantee accurate and reliable results, at quick turnaround time! Please contact us If you are interested in our services.
References
For research use only, not intended for any clinical use.