Edman Based Protein Sequencing
- Home
- Services
- Protein Sequencing
- Edman Based Protein Sequencing
Service Details
Edman degradation sequencing, allows for the tagging and cleavage of peptides from the N-terminus without breaking peptide bonds between other amino acid, is a mature protein sequencing method. With the development of mass spectrometry technology, the use of Edman degradation sequencing began to decrease, but Edman degradation sequencing remains a powerful and irreplaceable method for protein N-terminal sequencing. As an established sequencing method, Edman Degradation sequencing provides more accurate protein sequence data than MS. Which Could be used to verify the N-terminal boundaries of recombinant proteins or to determine the N-terminus of protease resistance domains, especially when the protein or domain is >40 - 80 kda or is not easily purified. In addition, some new proteins and peptides for which sequence databases are not available for MS/MS database searches can be analyzed using Edman degradation.
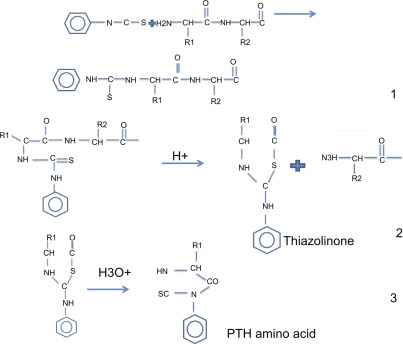 Fig. 1. The Edman degradation reactions and conversion step. (Khalid Z M, et al., 2021)
Fig. 1. The Edman degradation reactions and conversion step. (Khalid Z M, et al., 2021)
Creative Proteomics relies on the company's existing Edman sequencing system to provide N-terminal sequencing services for purified protein products, antibodies and protein vaccines for scientific researchers and scientific research customers. Using our sequencing system, the sequence information of the N-terminal 30 amino acids can be determined, while using a specific protein loading system, the N-terminal 60-70 amino acids can be determined. In addition, our professional protein sequencing platform also provides an N-terminal sequence analysis platform based on mass spectrometry, which can detect blocked and modified protein ends. The advantages of the two complement each other to ensure the smooth progress of N-terminal sequencing. The Edman based protein sequencing we provide is also suitable for the folowing services.
In order for you have a clearer understanding about our Edman sequencing services, Creative Proteomics has customized the following complete sequencing process for your protein.
Ⅰ. Under alkaline conditions, PITC binds to the free amino group at the N-terminus of the protein.
Ⅱ. In acidic solution, the N-terminal residue is cleaved.
Ⅲ. The PITC-bound residues were converted into more stable PTH residues.
Ⅳ. The amino acid species was determined by on-line HPLC analysis according to the elution time.
1. Liquid Samples
2. Electroblotted Samples
Creative Proteomics provides comprehensive Edman degradation sequencing projects for global customers. But if you have any special requirement, you are welcome to contact us for custom service. We look forward to your happy cooperation.
Question: Is there a requirement for protein sample purity in N-terminal sequencing?
Answer: Yes, ideally, the purity should be above 95%; otherwise, with multiple amino acids eluting in each cycle, it becomes challenging to attribute them to the protein sequence.
Question: If a protein has two or three chains, how can N-terminal sequencing be conducted?
Answer: Firstly, perform SDS-PAGE electrophoresis to separate the bands. Then transfer the protein from the gel to a PVDF membrane. After Coomassie Brilliant Blue staining, cut the corresponding bands for sequencing.
Two points to note: Avoid using Tris-glycine buffer during transfer, as it can cause higher background in N-terminal sequencing. CAPS buffer is recommended. Also, when staining the PVDF membrane, use Ponceau S instead of Coomassie Brilliant Blue.
Question: What kind of samples are not suitable for N-terminal sequencing using the Edman degradation method to analyze the sequence?
Answer: First, samples with blocked N-termini, as the reaction cannot proceed when the α-amino group at the N-terminus is modified. Second, samples with too many non-standard amino acids in the targeted N-terminal sequence. In such cases, where there are no corresponding standards, N-terminal sequencing is not feasible. Mass spectrometry can be used for sequence confirmation in these situations.
The study focuses on exploring the venom of the rear-fanged cat-eyed snake M. colubrinus, aiming to identify and characterize a peptide with inhibitory effects on muscle-type nicotinic acetylcholine receptors (nAChRs). This research contributes to understanding the diversity of snake venom components and their potential as sources for bioactive peptides.
Venom extracted from M. colubrinus specimens kept in the serpentarium.
Membranes of Torpedo californica used for competitive binding experiments.
ALEXA 488-labeled α-Bgt employed as a marker for nAChR binding.
1. Venom Extraction and Peptide Isolation:
2. Amino Acid Sequencing:
3. Peptide Synthesis:
4. Biological Activity of Macoluxin:
The venom of M. colubrinus inhibits nAChR, marking the first identification of such activity in a rear-fanged snake.
Macoluxin, a peptide from M. colubrinus venom, competitively inhibits muscle-type nAChR.
Amino acid sequence analysis reveals macoluxin's high similarity to a fragment of snake venom metalloproteinases.
Macoluxin's inhibitory effect on nAChR is demonstrated through competitive binding assays and electrophysiological experiments.
The study suggests that venom proteins can serve as precursors for diverse peptide toxins with distinct biological activities.
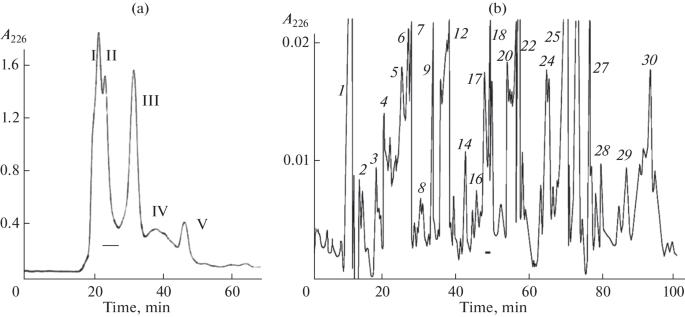 The isolation of active compound from the venom of M. colubrinus
The isolation of active compound from the venom of M. colubrinus
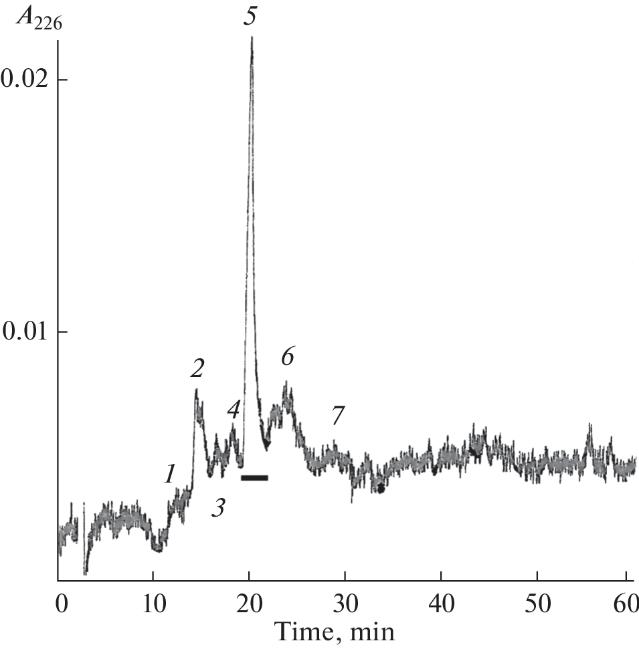
The separation of fraction 18 by reversed-phase chromatography on a Jupiter C18 column (4. 6 × 250 mm) in an acetonitrile concentration gradient of 20–30% for 60 min at a flow rate of 1 mL/min. The horizontal line indicates the active fraction.
Reference
For research use only, not intended for any clinical use.