Mass Spectrometry Based Protein Sequencing
- Home
- Services
- Protein Sequencing
- Mass Spectrometry Based Protein Sequencing
Service Details
Mass spectrometry based protein sequencing analysis is the analysis of the amino acid sequence of a sample (protein, antibody, peptide). Mass spectrometry for peptide and protein sequence determination includes bottom-up proteomics and top-down proteomics. The former is the mainstream technology of mass spectrometry sequencing, i.e., the protein is cleaved into small fragments by specific enzymatic or chemical hydrolysis, and then the molecular weight of each product peptide is detected by mass spectrometry, and the obtained peptide spectrum data is entered into a database to search for known proteins corresponding to it to obtain the protein sequence to be measured. This method can effectively identify amino acid isomers and post-translational modifications. Due to the introduction of cleavage, it is difficult to resolve and grasp the overall sequence information of the protein using this method, and it is difficult to identify proteins with low content. The latter method starts from the whole protein molecule, which can be introduced into the mass spectrometry by electrospray or matrix-assisted laser-resolved ionization, and then dissociated and analyzed by tandem mass spectrometry.
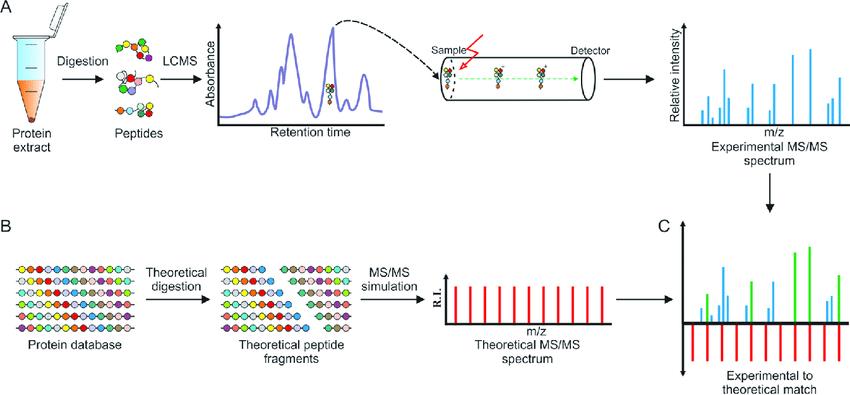
 Fig. 1. Protein sequencing based on the mass spectrometry. (Chait B T, 2006)
Fig. 1. Protein sequencing based on the mass spectrometry. (Chait B T, 2006)
At Creative Proteomics, we pride ourselves on offering a comprehensive range of protein sequencing services to meet the diverse needs of our clients. Our expertise and advanced technologies enable us to deliver accurate and reliable results for a variety of sequencing projects.
Protein N-Terminal Sequencing:

Protein C-Terminal Sequencing:
Protein Full-Length Sequencing:

Protein De Novo Sequencing and Mutation Analysis:
Top-Down Method Based Protein Sequencing:
Multiple Protease Digestion Platform: For enzymatic digestion, we employ a diverse set of proteases beyond the traditional Trypsin. This includes Chymotrypsin, Asp-N, Glu-C, Lys-C, and Lys-N, which are specifically selected based on the sample and the experimental objectives. This strategy enhances peptide sequence coverage and facilitates the analysis of complex samples.
Advanced Fragmentation Techniques: To ensure comprehensive coverage and to accurately identify post-translational modifications, we utilize both Higher-energy Collisional Dissociation (HCD) for generating b and y ions and Electron Transfer Dissociation (ETD) for preserving labile post-translational modifications. These techniques, available on our Orbitrap systems, provide complementary information that enhances sequence coverage and modification detection.
High-Resolution Mass Spectrometers: Our suite includes the Thermo Scientific™ Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer and the Bruker Daltonics timsTOF Pro. These instruments are renowned for their exceptional resolution, mass accuracy, and sensitivity, enabling the precise measurement of peptide masses and the identification of proteins even at low abundances.
Integrated Data Analysis Software: For data analysis, we employ sophisticated software solutions such as the Mascot search engine and Proteome Discoverer™ for protein identification and quantitation. These tools are critical for interpreting the complex data generated by mass spectrometry and for ensuring the accuracy of our protein sequencing efforts.
 Peptide identification in mass-spectrometry shotgun approach (Chugunovae et al, 2018).
Peptide identification in mass-spectrometry shotgun approach (Chugunovae et al, 2018).
![]() Instrumentation: We employ cutting-edge mass spectrometry platforms with resolution exceeding 100,000 and mass accuracy within 1 ppm, ensuring the highest level of precision and sensitivity.
Instrumentation: We employ cutting-edge mass spectrometry platforms with resolution exceeding 100,000 and mass accuracy within 1 ppm, ensuring the highest level of precision and sensitivity.
![]() Throughput: Our instruments are capable of processing up to 500 samples per day, enabling rapid data generation without compromising quality.
Throughput: Our instruments are capable of processing up to 500 samples per day, enabling rapid data generation without compromising quality.
![]() Quality Metrics: We adhere to rigorous quality control measures, with a data accuracy rate exceeding 99.9% and a reproducibility rate of 98%, ensuring reliable and consistent results.
Quality Metrics: We adhere to rigorous quality control measures, with a data accuracy rate exceeding 99.9% and a reproducibility rate of 98%, ensuring reliable and consistent results.
![]() Data Interpretation: Our team of bioinformatics experts provides comprehensive data interpretation, delivering detailed insights and actionable conclusions.
Data Interpretation: Our team of bioinformatics experts provides comprehensive data interpretation, delivering detailed insights and actionable conclusions.
![]() Publication Record: Our work has resulted in over 500 peer-reviewed publications in prestigious journals, underscoring our expertise and contributions to the field.
Publication Record: Our work has resulted in over 500 peer-reviewed publications in prestigious journals, underscoring our expertise and contributions to the field.
![]() Tailored Workflows: We develop customized workflows tailored to each project's unique requirements, resulting in optimized efficiency and cost-effectiveness.
Tailored Workflows: We develop customized workflows tailored to each project's unique requirements, resulting in optimized efficiency and cost-effectiveness.
| Sample Type | Volume/Amount | Preparation Guidelines |
|---|---|---|
| Pure Proteins | 10-100 µg | - Lyophilized or in solution - Purity > 90% - Avoid detergents and contaminants |
| Complex Mixtures | 100 µg - 1 mg | - Precipitation and digestion - Removal of salts and detergents |
| FFPE Samples | 10 sections (10 µm thick) | - Deparaffinization - Protein extraction and digestion |
| Cell Lysates | 100 µg - 2 mg | - Lysis buffer without detergents - Centrifugation to remove debris |
| Tissue Samples | 10-100 mg | - Homogenization - Extraction buffer without detergents |
![]() Protein Identification: Unambiguously identifying proteins in complex mixtures, crucial for understanding cellular processes and disease states.
Protein Identification: Unambiguously identifying proteins in complex mixtures, crucial for understanding cellular processes and disease states.
![]() Post-Translational Modifications (PTMs) Analysis: Detecting and characterizing PTMs, essential for grasping protein function and regulation.
Post-Translational Modifications (PTMs) Analysis: Detecting and characterizing PTMs, essential for grasping protein function and regulation.
![]() Protein Quantification: Quantitative proteomics, providing insights into differential expression patterns under various conditions.
Protein Quantification: Quantitative proteomics, providing insights into differential expression patterns under various conditions.
![]() Protein-Protein Interactions: Elucidating the interaction networks, pivotal for deciphering signaling pathways and functional dynamics.
Protein-Protein Interactions: Elucidating the interaction networks, pivotal for deciphering signaling pathways and functional dynamics.
![]() Drug Discovery: Target identification and validation, accelerating the development of therapeutic agents.
Drug Discovery: Target identification and validation, accelerating the development of therapeutic agents.
![]() Biomarker Discovery: Identifying disease-specific proteins, enhancing diagnostic and prognostic capabilities.
Biomarker Discovery: Identifying disease-specific proteins, enhancing diagnostic and prognostic capabilities.
Reference
1. Can the bands on SDS gels be cut and sent for mass spectrometry sequencing?
Yes, bands separated by SDS can be directly cut and sent for mass spectrometry identification. To enhance the accuracy of identification, it's important to use an appropriate gel concentration for separating the target protein bands and try to separate neighboring protein bands from the target protein as much as possible during gel electrophoresis.
2. Can protein bands on SDS gels that are too thin be identified?
Generally, for protein bands stained with Coomassie or SYPRO Ruby, bands visible to the naked eye are sufficient for sequencing.
3. What if the cut SDS protein bands contain more than one band?
If the cut protein bands contain multiple bands, they can still be analyzed for protein identification. Subsequently, protein sequences can be inferred through data comparison. In fact, most protein bands cut from SDS-PAGE gels contain multiple proteins. Analyzing these complex proteins can be achieved through chromatography-mass spectrometry tandem identification methods, where complex proteins are first separated by chromatography and then analyzed one by one through mass spectrometry, achieving higher accuracy.
4. Can Western blot results be used for protein sequencing?
Proteins transferred after Western blotting can indeed be sequenced. By comparing Western blotting results to SDS-PAGE gels, the corresponding positions of protein bands on SDS-PAGE gels can be identified and then cut for mass spectrometry identification. If the protein quantity is sufficient and the purity is high, protein bands on PVDF membranes can be directly cut for N-terminal sequencing identification.
5. What precautions should be taken during protein sample preparation?
Mass spectrometry for protein identification is highly sensitive, and even trace amounts of contaminating proteins introduced during the operation can be detected, significantly affecting the accuracy of protein sequencing analysis. Therefore, during sample preparation for mass spectrometry analysis, clean and uncontaminated vessels, reagents meeting the level of purity required for mass spectrometry identification, freshly prepared solutions, gloves, and head covers should be used to avoid contamination by keratin and other contaminants.
6. What are the requirements for preparing various types of samples for protein sequencing?
In general, protein samples separated by SDS-PAGE gels and stained with Coomassie or SYPRO Ruby are compatible with mass spectrometry identification. However, protein samples stained with silver must not use glutaraldehyde as a fixative, as it would affect subsequent mass spectrometry analysis. Additionally, when the protein concentration in the solution is low, the use of surfactants like SDS should be minimized, and attention should be paid to reducing salt concentration to improve the accuracy of protein identification.
7. How to prepare protein samples if the molecular weight of the protein of interest is small?
If the molecular weight of the protein of interest is relatively small, high-concentration SDS-PAGE gels can be used to separate proteins, followed by staining with Coomassie, and then cutting bands of protein gel that are consistent in size with the target protein for mass spectrometry identification.
8. What are the requirements for sample shipment?
Protein bands and powder are relatively stable and can be transported using ice packs. Protein solution should be shipped using dry ice. It is recommended to lyophilize the samples before shipping, as proteins are highly stable in lyophilized form. Care should be taken to avoid repeated freeze-thaw cycles to prevent protein degradation.
9. Why is it necessary to enzymatically digest proteins into peptides before mass spectrometry sequencing?
The larger the protein fragments, the lower the accuracy of mass spectrometry detection. Therefore, before mass spectrometry detection, proteins need to be digested into smaller peptides to improve the accuracy of mass spectrometry detection. Generally, peptides with 6-20 amino acids are most suitable for mass spectrometry detection.
10. How to improve the efficiency of determining the sequence of modified peptide segments in protein sequencing?
Under current conditions, it is difficult for mass spectrometry to provide 100% sequence coverage of peptides. Some peptides may be lost during the process, and protein phosphorylation can inhibit trypsin digestion. Moreover, the content of phosphorylated peptides is much lower than that of non-phosphorylated peptides, which may lead to inhibition of mass spectrometry response. Therefore, efforts should be made to minimize the content of non-phosphorylated peptides. Methods such as fractionation, IMAC (immobilized metal affinity chromatography), and antibody binding can be used. MALDI-TOF-MS can be used to determine the molecular weight of peptide segments. If the obtained molecular weight of a peptide segment is 80 Da or its multiples larger than the expected molecular weight, the presence of phosphorylation modification can be determined.
11. What could cause two amino acid residues to undergo Edman degradation simultaneously in the determination of protein N-terminal amino acid sequences?
When two amino acid residues are detected simultaneously in Edman degradation, it is possible that the protein is impure and contaminated with other proteins. If one of the amino acids is glycine, it is possible that residual glycine from the buffer in the protein band was not completely removed, leading to this result.
For research use only, not intended for any clinical use.