Introduction of Protein Sequencing
Protein sequencing is the process of determining the amino acid sequence of all or part of the primary structure of a protein, which determines the advanced structure of the protein and affects the function of the protein in. The determination of the amino acid order of proteins is the basis of protein chemistry research. A large amount of information can be obtained by analyzing the amino acid sequence of a protein, and which can be applied to other related fields, such as protein identification, design of molecular cloning probes and the development of biological drugs. Since the determination of the primary structure of insulin by F. Sanger in 1953, the primary structures of about 100,000 different proteins are now known.
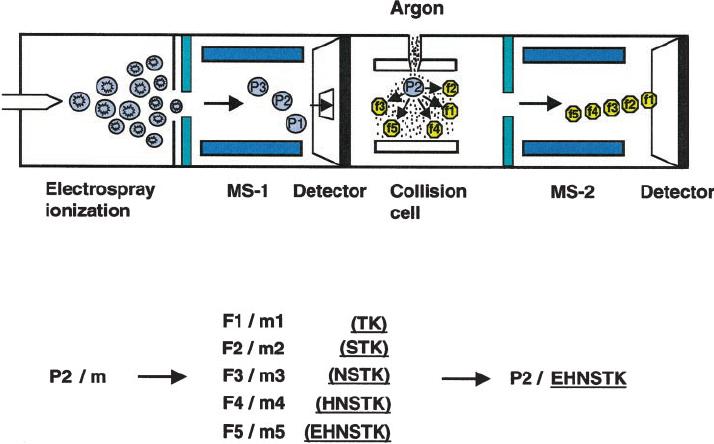 Schematic of peptide sequencing by tandem mass spectrometry (Li et al., 2000).
Schematic of peptide sequencing by tandem mass spectrometry (Li et al., 2000).
Protein Sequencing Service in Creative Proteomics
Creative Proteomics provides the two main methods, are Edman degradation and mass spectrometry (MS) analysis, that are commonly used for protein sequencing, of which mass spectrometry being the most widely used method for protein sequencing. For proteins with unknown theoretical sequences, we provide protein de novo sequencing services to analyze protein sequences. In addition, our comprehensive protein sequencing services are also widely used for the researches of protein identification, PTM analysis, glycoproteomics, peptidomics and so on.
Creative Proteomics offers the following featured protein sequencing analysis services.
Utilizing advanced mass spectrometry, we provide accurate sequencing and comprehensive characterization of proteins, including post-translational modifications.
Reliable Edman degradation methodology for precise amino acid sequencing, facilitating primary structure determination essential for biotechnology and pharmaceutical applications.
We employ nanopore technology to precisely identify amino acids and differentiate short peptides, offering real-time analysis with high sensitivity.
With single molecule fluorescence sequencing, we detect and sequence proteins, offering high sensitivity for analyzing small protein samples and providing insights into protein structure and function.
Advantages of Protein Sequencing Services
- Expertise: With over 20 years in the field, our team's expertise ensures accurate, reliable results across a broad spectrum of protein types and complexities.
- Advanced Technology: Utilizing cutting-edge LC-MS/MS and Edman Degradation technologies, we offer unparalleled resolution in protein sequencing.
- Comprehensive Coverage: Our ability to provide full-length sequencing, including detailed analysis of PTMs and termini, sets us apart, offering a holistic view of protein structures.
- Customized Solutions: Understanding that each project has unique requirements, we tailor our services to meet the specific needs of our clients, ensuring optimal outcomes.
Protein Sequencing Applications
Protein sequencing is an indispensable tool across various scientific fields, offering deep insights into biological processes and advancing research and development projects. At Creative Proteomics, we specialize in providing precise and comprehensive protein sequencing services tailored to meet the needs of diverse applications:
Drug Discovery and Biomarker Development: Identifying drug targets and developing biomarkers for disease diagnosis and treatment monitoring.
Biosimilars and Biopharmaceutical Development: Ensuring biosimilarity in biopharmaceuticals through detailed amino acid sequence analysis, critical for regulatory approval.
Protein Function and Interaction Studies: Mapping protein interactions and functions to elucidate biological pathways and mechanisms.
Structural Biology and Protein Engineering: Determining protein structures and engineering proteins with specific characteristics for new therapeutics and industrial enzymes.
Quality Control in Manufacturing: Serving as a quality control measure in the production of therapeutic proteins and antibodies.
Evolutionary Biology and Systematics: Tracing evolutionary relationships and understanding protein function conservation across species.
Agricultural Biotechnology: Enhancing crop disease resistance, quality, and yield through protein engineering.
Environmental Proteomics: Studying organism adaptations to environmental stressors and understanding microbial roles in biogeochemical cycles.
Neuroscience: Unraveling protein interactions in the brain to inform on neurological disease mechanisms and therapies.
Creative Proteomics focuses on the development and research of protein sequencing platforms. We have a professional scientific research team who dedicated to providing the most professional and economical protein sequencing service projects for our global researchers. If you are interested in our services or have questions about your protein sequencing, please feel free contact us. We look forward to working with you.
Reference
- Li, Jiaxu, and Sarah M. Assmann. "Mass spectrometry. An essential tool in proteome analysis." Plant physiology 123.3 (2000): 807-810.
Case: Biochemical Characterization of Tauopathies Reveals Distinct Core Units in Pathological Tau Aggregates
Background
Tauopathies, neurodegenerative disorders marked by abnormal tau protein accumulation, exhibit diverse histopathologies. This study aims to biochemically classify tauopathies—Alzheimer's disease (AD), Pick's disease (PiD), corticobasal degeneration (CBD), progressive supranuclear palsy (PSP), and MAPT intronic mutations—by investigating sarkosyl-insoluble and trypsin-resistant tau fragments. The focus is on identifying disease-specific core units within tau aggregates and understanding potential prion-like mechanisms.
Samples
The study utilized sarkosyl-insoluble fractions extracted from the brains of individuals diagnosed with Alzheimer's disease (AD), Pick's disease (PiD), corticobasal degeneration (CBD), progressive supranuclear palsy (PSP), and cases with MAPT intronic mutations.
Technical Methods
1. Immunoblot Analysis:
- Employed the anti-tau antibody T46 for the detection of C-terminal tau fragments.
- Investigated hyperphosphorylated full-length tau and C-terminal tau fragments.
- Identified characteristic band patterns crucial for the biochemical diagnosis of tauopathies.
2. In-Gel Digestion and LC/MS/MS Analysis:
- Attempted to unravel the N-terminal sequences of tau fragments.
- Revealed heterogeneous N-termini, indicating a diversity in conformation.
3. Trypsin Treatment and Immunoblotting:
- Explored protease-resistant tau fragments using trypsin.
- Utilized the TauC4 antibody to detect trypsin-resistant cores.
- Observed distinct trypsin-resistant band patterns specific to each tauopathy.
4. MALDI-TOF MS Analysis:
- Analyzed trypsin-resistant tau fragments from AD brains.
- Identified molecular masses corresponding to specific tau isoforms.
- Detected signals suggesting phosphorylation.
5. Electron Microscopy:
- Applied immunoelectron microscopy to investigate the correlation between trypsin-resistant cores and the morphology of tau fibrils.
- Observed distinct tau fibril structures in various tauopathies.
Results
Identified disease-specific core units of tau aggregates positioned between residues 243–406.
Biochemically classified tauopathies into at least four types: PiD, PSP, CBD, and AD.
Discerned distinct banding patterns of C-terminal fragments and trypsin-resistant tau among different tauopathies.
Noted similarities to prion diseases in terms of proteinase-K-resistant banding patterns.
Proposed models of tau fibril cores based on an antiparallel association, suggesting the involvement of various homodimers and heterodimers in fibril assembly.
 N-terminal sequences identified by protein sequencing, C-terminal peptides identified by LC/MS/MS analysis of the V8 digests, or predicted sequences by MALDI-TOF MS analysis.
N-terminal sequences identified by protein sequencing, C-terminal peptides identified by LC/MS/MS analysis of the V8 digests, or predicted sequences by MALDI-TOF MS analysis.
Reference
- Taniguchi-Watanabe, Sayuri, et al. "Biochemical classification of tauopathies by immunoblot, protein sequence and mass spectrometric analyses of sarkosyl-insoluble and trypsin-resistant tau." Acta neuropathologica 131 (2016): 267-280.

 Schematic of peptide sequencing by tandem mass spectrometry (Li et al., 2000).
Schematic of peptide sequencing by tandem mass spectrometry (Li et al., 2000).
 N-terminal sequences identified by protein sequencing, C-terminal peptides identified by LC/MS/MS analysis of the V8 digests, or predicted sequences by MALDI-TOF MS analysis.
N-terminal sequences identified by protein sequencing, C-terminal peptides identified by LC/MS/MS analysis of the V8 digests, or predicted sequences by MALDI-TOF MS analysis.