Protein Degradation or Enzyme Digestion Analysis
- Home
- Services
- Protein Sequencing
- Edman Based Protein Sequencing
- Protein Degradation or Enzyme Digestion Analysis
Service Details
Edman degradation is a time-honored N-terminal sequencing technique for proteins and cleavage fragments. Proteolytic processing modifies the pleiotropic functions of many large, complex, and also reveal cryptic binding sites and generate novel or modified biologically active cleavage products displaying new N- and C-termini. Proteolytic processing is a ubiquitous and irreversible post-translational modification involving limited and highly specific hydrolysis of peptide and isopeptide bonds of proteins by proteases. The identification of exact proteolytic cleavage sites in the extracellular matrix laminins and other extracellular matrix proteins is not only important for understanding protein turnover but is needed for the identification of new bioactive cleavage products. However, the identification of cleavage sites in complex macromolecular proteins such as those that make up the ECM is not trivial. N-terminal microsequencing of proteolytic fragments is a commonly used method to distinguish the N-terminus of a protein from the N-terminus of its protease cleavage product will accelerate the identification of protease substrates with broad or unknown specificity.
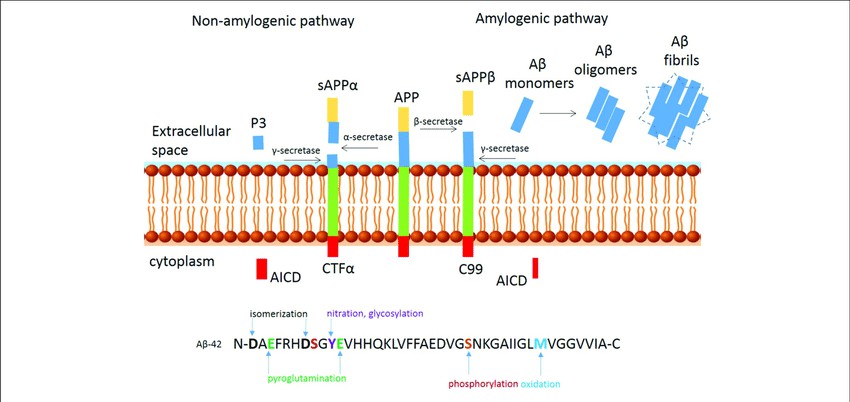 Fig. 1. Proteolytic processing of amyloid
precursor protein. (Sumner I E, et al., 2018)
Fig. 1. Proteolytic processing of amyloid
precursor protein. (Sumner I E, et al., 2018)
Over the past few decades, Creative Proteomics has focused on developing powerful protein sequencing platforms for the characterization of proteolysates and native protein N/C termini. At Creative Proteomics, our scientists prefer Edman degradation to determine your cleavage sites and analyze the N-terminus of new protein fragments formed by degradation or enzymatic cleavage. It is an important tool for many scientific areas including:
Our sequencing method, Edman degradation, is a procedure that requires a free N-terminal amino group on the peptide or protein. The cyclical process involves the coupling of sequencing reagent, phenyl isothiocyanate (PITC), to the free amino group of the N-terminal amino acid which is then selectively cleaved. This PITC coupled residue is converted to a stable PTH-residue and after separation by HPLC chromatography, the amino acid can be identified. The Edman reaction cycle is repeated for the number of cycles you require until a sequence of amino acids is determined.
At Creative Proteomics, our scientists developed a new liquid chromatography-mass spectrometry (LC-MS) method to complement N-terminal Edman sequencing for N-terminal identification of protein cleavage fragments in solution. Because N-terminal Edman sequencing of multiple and often closely spaced cleaved fragments on an SDS-PAGE gel is difficult, limiting throughput and coverage. Our LC-MS method can identify hundreds of peptides in a single analysis, making it ideal for identifying multiple cleavage sites in a single protein.
In addition to a clear and easy-to-understand report, you can expect:
Creative Proteomics provides global customers with professional protein degradation or enzyme digestion analysis service to analyze the N-terminus of the new protein fragment formed by degradation or enzyme cleavage and thus to determine the cleavage site. At Creative Proteomics, our highly qualified team works with our customers stand together on the front lines to help you solve tough research challenges. If you are interested in our services, please contact us immediately.
References
For research use only, not intended for any clinical use.