Introduction
Sequence variants are erroneous amino acid substitutions in the primary structure of proteins. During therapeutic
protein development, sequence variants may cause protein misfolding, aggregation, and eventually affect drug safety
and efficacy; thus, sequence variant analysis (SVA) is of great importance in product and process development.
Depending on the origin, sequence variants can be classified into 2 categories, mutations and misincorporations.
Mutations are DNA-level errors that are most commonly introduced by incorrect nucleotide incorporations during DNA
replications. In contrast, misincorporations are due to incorrect mRNA or amino acid incorporations during
transcription (DNA to mRNA) or translation (mRNA to protein) stages. While mutations can be detected at both DNA and
protein levels, amino acid misincorporation (sequence variants) can only be detected at the protein level. Tandem
mass spectrometry (LC-MS/MS) is the most frequently used tool to detect sequence variants at the protein level.
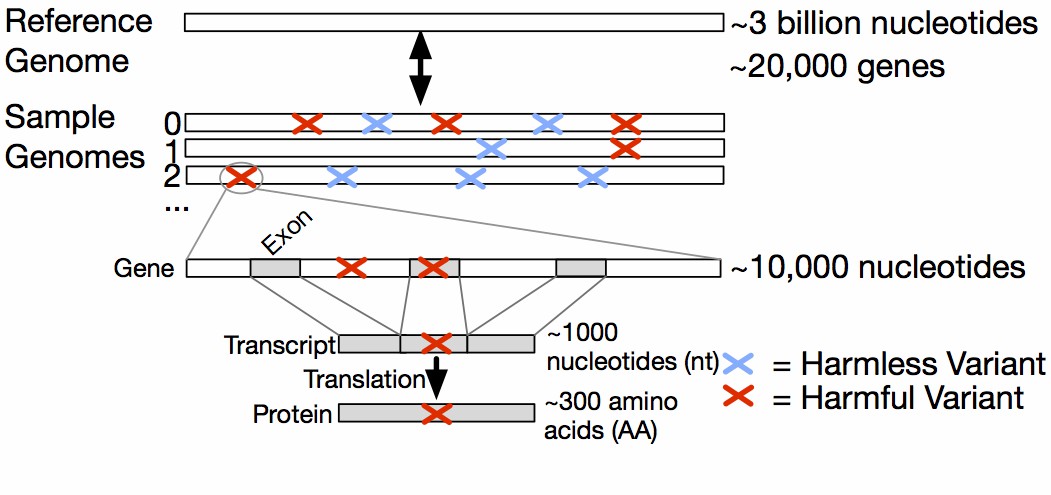 Fig.
1. Biological context in which variants occur. The gene level is specified by genomic coordinates, the
exon-containing transcript level is specified by transcript coordinates, and the protein level is specified by
protein coordinates. (Ferstay J A, et al., 2013)
Fig.
1. Biological context in which variants occur. The gene level is specified by genomic coordinates, the
exon-containing transcript level is specified by transcript coordinates, and the protein level is specified by
protein coordinates. (Ferstay J A, et al., 2013)
Our Services
Creative Proteomics has developed a complete set of unknown protein/peptide sequence and mutation
analysis services based on the de novo sequencing platform combined with bioinformatics tools, which can accurately
and completely determine the complete sequence and all mutations in protein products, even in the case that protein
is heavily mutated. The sequence variation platform we developed that enables them to identify all amino acid
sequence variation in a mixture by combining HPLC-UV/MS/MS characterization of peptide maps generated using multiple
protease digestions with bioinformatics' SIEVE analysis software, and this method can easily detected the
sequence variant.
Our goal is to provide high-quality sequence variation services to our global customers and to support the
development of therapeutic protein development. We have carried out SVA screening or several dozen products for
global customer, and our screening cover the entire mAb/protein sequence. With our technology, we can detect heavily
mutated proteins, and we can detect very low-level mutations due to translational errors.
Our Technical Advantages
- 100% sequence coverage based on multiple digestions of different enzymes.
- Capable of detecting mutations at as low as 1%, some as low as 0.1% of the protein level.
- Capable of obtaining a semi-quantitative measurement of a mutation level.
Applications
- Analysis of the exact amino acid sequence of commercially available protein products with high commercial value,
where the amino acid sequence has been altered to provide better or new properties.
- Analysis of new proteins, of which the amino acid sequence is completely unknown.
- Unwanted mutations are screened at the protein level during clonal selection prior to production of protein
therapeutics.
- Sequence variant identification in recombinant biopharmaceuticals.
Our sequence and mutation analysis of unknown proteins or peptides services are widely used in the development of
protein therapy drugs. At Creative Proteomics, highly skilled and dedicated scientific staff
ensures that the most appropriate methods and technologies are selected for you. If you have any special
requirements about our services, please feel free to contact us. We are
looking forward to working together with you.
References
- Li W, Wypych J, Duff R J. (2017) Improved sequence variant analysis strategy by automated false positive
removal. MAbs. 9(6):978-984.

 Fig.
1. Biological context in which variants occur. The gene level is specified by genomic coordinates, the
exon-containing transcript level is specified by transcript coordinates, and the protein level is specified by
protein coordinates. (Ferstay J A, et al., 2013)
Fig.
1. Biological context in which variants occur. The gene level is specified by genomic coordinates, the
exon-containing transcript level is specified by transcript coordinates, and the protein level is specified by
protein coordinates. (Ferstay J A, et al., 2013)