Introduction
The development of stable cell lines for biotherapeutics is an irreversible process, and Sequence variants (SVs)
resulting from unintended amino acid substitutions in recombinant therapeutic proteins have increasingly gained
attention from the biopharmaceutical industry given their potential impact on efficacy and safety. For
well-optimized production systems, such sequence variants are usually present at very low levels in the final
protein product due to the high fidelity of DNA replication and protein biosynthesis processes in mammalian
expression systems. However, their levels can be significantly elevated in cases where the selected production cell
line is not fully optimized of the manufacturing process or has unexpected DNA mutations. Small amounts of SV of the
protein products are prone to occur during cell line construction and storage, which may also cause changes in
biological activity and generate human immune responses, thereby creating a huge potential risk. Therefore, it is
very important to detect this natural mutation before large-scale production. Currently, mass spectrometry (MS) The
application of advanced methods enables the highly sensitive detection of sequence variants (SV), such as the
characterization of cell lines and recombinant proteins.
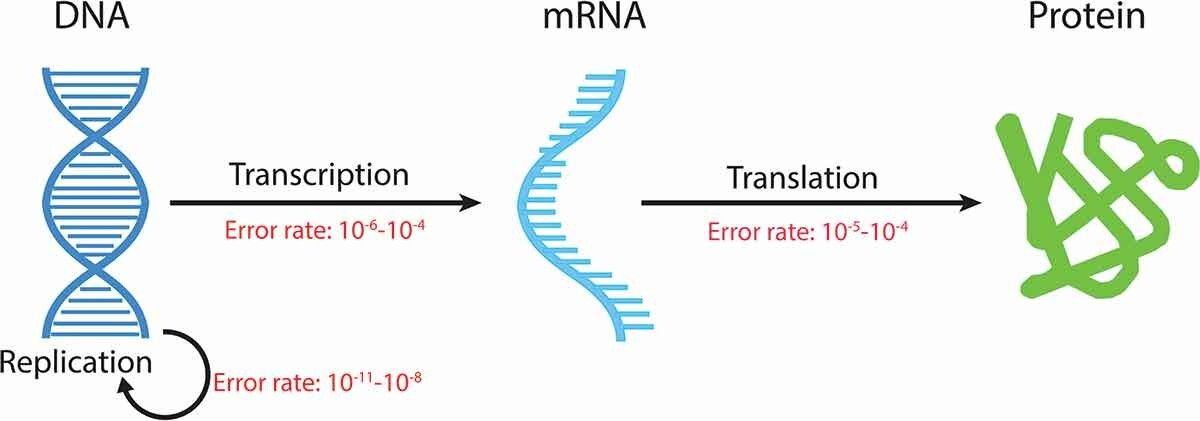 Fig. 1. Elucidation of the
central dogma and typical error rate in each step. (Zhang A M, et al., 2020)
Fig. 1. Elucidation of the
central dogma and typical error rate in each step. (Zhang A M, et al., 2020)
Our Services
Creative Proteomics has developed a complete set of mutation analysis services based on the de novo
sequencing platform combined with bioinformatics tools, which can accurately and completely determine all mutations
in protein products, even in the case that protein is heavily mutated. The sequence variation platform we developed
that enables them to identify all amino acid sequence variation in a mixture by combining HPLC-UV/MS/MS
characterization of peptide maps generated using multiple protease digestions with bioinformatics' SIEVE
analysis software, and this method can easily detected the sequence variant. With our technology, we can detect
heavily mutated proteins, and we can detect very low-level mutations due to translational errors. The methods we
have developed that can detect sequence differences as low as 1 amino acid and can cover the whole of
protein/antibody.
Our Technical Advantages
- 100% sequence coverage based on multiple digestions of different enzymes.
- Capable of detecting mutations at as low as 1%, some as low as 0.1% of the protein level.
- Capable of obtaining a semi-quantitative measurement of a mutation level.
Our sequence mutation aanalysis of proteins expressed in stable cell Lines services are widely used in the
development of protein therapy drugs. At Creative Proteomics, highly skilled and dedicated
scientific staff ensures that the most appropriate methods and technologies are selected for you. Please feel free
to contact us if you have any special requirements about our services. We
are looking forward to working with you.
References
- Zhang A M, Chen Z W, Li M N, et al. (2020) A general evidence-based sequence variant control limit for
recombinant therapeutic protein development. MAbs. 12(1):1791399.
- Li W, Wypych J, Duff R J. (2017) Improved sequence variant analysis strategy by automated false positive
removal. MAbs. 9(6):978-984.

 Fig. 1. Elucidation of the
central dogma and typical error rate in each step. (Zhang A M, et al., 2020)
Fig. 1. Elucidation of the
central dogma and typical error rate in each step. (Zhang A M, et al., 2020)