Protein Mass Spectrometry Identification Service
- Home
- Applications
- Proteomics Analysis Services
- Protein Identification Services
- Protein Mass Spectrometry Identification Service
Service Details
Mass spectrometry (MS) is a commonly used, high-throughput tool for studying proteins. The procedure of MS-based protein identification involves digesting proteins into peptides, which are then separated, fragmented, ionised, and captured by mass spectrometers. Proteins are finally identified from the peaks of the captured mass spectra using computational methods, where each peak theoretically represents a peptide fragment ion. Existing Mass spectrometry based protein identification methods can be categorised into 2 approaches: the database search approach and the de novo sequencing approach. The database search approach identifies proteins by generating theoretical spectra in silico from a given protein sequence database and comparing experimental spectra with the theoretical ones to find the closest matches. The de novo sequencing approach identifies proteins by extracting protein sequence information directly from the spectrum peaks derived from peptide fragment ions without recourse to any protein database.
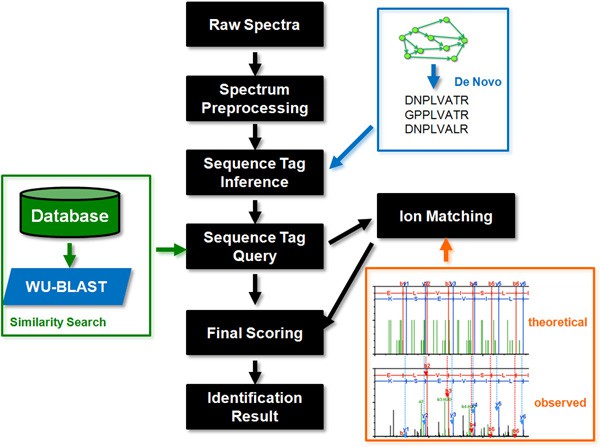 Fig.
1. Brief overview of the NovoDB identification method. (Wang P H, et al., 2013)
Fig.
1. Brief overview of the NovoDB identification method. (Wang P H, et al., 2013)
Mass spectrometry-based protein identification is a very challenging task. Creative Proteomics has developed a new integrative protein identification method which can integrate de novo sequencing more efficiently into database searching. Our scientists firstly infer partial peptide sequences, known as tags, directly from tandem spectra through de novo sequencing, and then puts these sequences into a database search to find a close peptide match.
At Creative Proteomics, we developed the following complete set of protein identification workflows, including spectrum preprocessing, sequence tag inference, sequence tag query, spectrum ion match, and final scoring.
Ⅰ. Our scientists use two versions of the peak intensities to preprocess the spectra and normalise the peak intensities, and peak into five regions based on the mass to charge ratio and utilises this information in the sequence tag inference.
Ⅱ. A number of peptide sequence tags directly are infered from the spectrum according to the above spectrum information.
Ⅲ. After sequence tags are obtained, the next stage is to query a database to find see if matches can be found.Spectrum ion match.
Ⅳ. Then based on the dot-product between the observed ions were matched with the theoretical ions generated in silico from the sequence database.
Ⅴ. The target protein is identified according to the final score calculated by the bioinformatics software analysis, and a corresponding report is given.
Thanks to our powerful mass spectrometry sequencing platform, Creative Proteomics, as a first-class protein sequencing service provider, is committed to providing global customers with all protein mass spectrometry identification services to accelerate their project research journey. Our services guarantees accurate and reliable results, at quick turnaround time! Please contact us If you are interested in our services.
References
For research use only, not intended for any clinical use.