N-Terminal Sequencing (N-Terminal Unblocked)
- Home
- Services
- Protein Sequencing
- Edman Based Protein Sequencing
- N-Terminal Sequencing (N-Terminal Unblocked)
Service Details
Edman Degradation Sequencing, allows for the tagging and cleavage of peptides from the N-terminus without breaking peptide bonds between other amino acid, is a mature protein sequencing method. With the development of mass spectrometry technology, the use of Edman degradation sequencing began to decrease, but Edman degradation sequencing remains a powerful and irreplaceable method for protein N-terminal sequencing. As an established sequencing method, Edman degradation sequencing provides more accurate protein sequence data than MS. Which could be used to verify the N-terminal boundaries of recombinant proteins or to determine the N-terminus of protease resistance domains, especially when the protein or domain is >40 - 80 kda or is not easily purified. In addition, some new proteins and peptides for which sequence databases are not available for MS/MS database searches can be analyzed using Edman degradation.
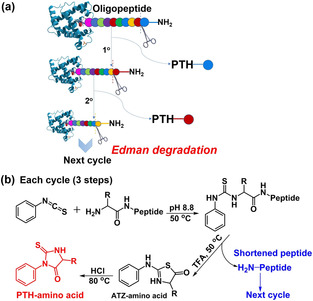 Fig. 1. (a) N-terminal Edman sequencing chemistry and (b) detailed procedure for each cycle. (Zhang H, et al., 2020)
Fig. 1. (a) N-terminal Edman sequencing chemistry and (b) detailed procedure for each cycle. (Zhang H, et al., 2020)
To elucidate the N-terminal amino acids of proteins, Creative Proteomics developed Edmand-based protein N-terminal sequencing technology relieing on the company's existing Edman sequencing system, a classical stepwise chemical cleavage method by automated phenyl isothiocyanate chemistry. The N-terminal amino acid group of the protein is labeled with phenyl isothiocyanate (PITC), the labeled N-terminal amino acid was selectively cleaved, and the extracted phenylthiohydantoin is identified by MS Urea (PTH)-amino acid, according to this procedure to determine the amino acid sequence of the protein. The specific workflow is as follows.
Ⅰ. Under alkaline conditions, PITC binds to the free amino group at the N-terminus of the protein.
Ⅱ. In acidic solution, the N-terminal residue is cleaved.
Ⅲ. The PITC-bound residues were converted into more stable PTH residues.
Ⅳ. The amino acid species was determined by on-line HPLC analysis according to the elution time.
At Creative Proteomics, Our N-terminal sequencing technology services can provide N-terminal sequencing analysis methods for biopharmaceutical proteins or other biological reagents such as Edman sequencing. The problems of N-terminal blocking and PTM restriction can be addressed by combining mass spectrometry, these two approaches complement each other to obtain comprehensive N-terminal sequence analysis information.
Creative Proteomics has professional protein N-terminal sequencing research resources. We provide you with high-quality one-stop N-terminal sequencing services with first-class experimental platforms and mature technical means. In particular, we will provide you with comprehensive GLP/cGMP compliant N-terminal sequencing analysis services around ICH guidelines (especially ICH Q6B) and the "Points to Consider" document issued by the US FDA. If you are interested in our services, please feel free to contact us.
References
For research use only, not intended for any clinical use.