Mass Spectrometry Data Processing and Analysis Service
- Home
- Applications
- Bioinformatics Services
- Mass Spectrometry Data Processing and Analysis Service
Service Details
Mass spectrometry (MS) is a powerful analytical technique that works by measuring the mass-to-charge ratio (m/z) of molecules present in a sample. The technique involves ionizing a sample and then stratifying the resulting ions according to their mass-to-charge ratio to generate large amounts of mass spectral data. Today, mass spectrometry, as a key analytical technique for protein research and biomolecular research in general, is driven by the need to identify, characterize and quantify proteins with ever-increasing sensitivity and in increasingly complex samples. New mass spectrometry-based analysis platforms and experimental strategies, allowing protein identification, annotation of secondary modifications, and determination of absolute or relative abundance of individual proteins. LC-MS-based methods have generated great interest for the analysis of complex peptide or protein mixtures due to their potential for full automation and high sampling rates.
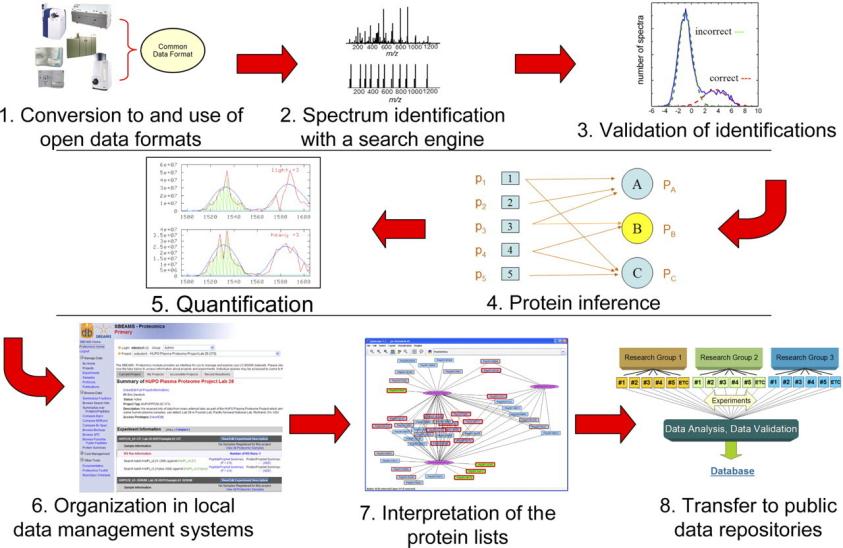 Fig.
1. Schematic overview of the typical workflow of the proteomics informatics processing of a data set. (Deutsch E W,
et al., 2008)
Fig.
1. Schematic overview of the typical workflow of the proteomics informatics processing of a data set. (Deutsch E W,
et al., 2008)
The continuous development of mass spectrometry technology is accompanied by the generation of rich mass spectrometry data. Although detailed mass spectrometry data information is conducive to more in-depth research on proteology and proteomics, the large amount of protein sample data generated by high-throughput proteomics detection also brings upgraded challenges to the analysis of protein sample characteristics. As a leading protein sequencing service company, Creative Proteomics has an advanced mass spectrometry analysis platform that is capable of comprehensive analysis of the mass spectrometry data generated.
Creative Proteomics' scientists have many years of experience in mass spectrometry. Here, we provide the following mass spectrometry data processing and analysis services, including but not limited to.
Our protein sequence analysis service is one of a wide range of analysis methods for protein or peptide sequences, with the purpose of studying their characteristics, function, structure or evolutionary process. And on this basis, our scientists can determine the function of the protein by studying the similarities between different sequences. Many tools and techniques have been chosen by Creative Proteomics to analyze alignment products and provide sequence comparisons to study their biology, including alignment-free sequence analysis, hidden markov model, regression analysis, support vector machine, artificial neural network method, bayesian network method, etc.
The PTMs of proteins allow the generation of many different protein species, thus compensating for the incongruity between the relatively few coding genes and the large number of variable regulators in complex organisms. Here, our expert team provides the numerous PTM data analysis services, including PTM predictive analysis, conserved sequence analysis of PTMs, and PTM crosstalk analysis.
Functional annotation and enrichment analysis have been widely used in bioinformatics for proteomics research. As one of the world's leading protein sequencing and mass spectrometry companies, we provide you with a wide range of functional annotation and enrichment analysis services, including statistical analysis of differential protein, GO functional annotation and enrichment analysis, COG functional annotation and enrichment analysis, differentially expressed protein cluster analysis, and protein Interaction network analysis, etc.
Creative Proteomics is a leading biotechnology company specializing in a range of protein analysis services. Our experienced scientists provide our clients with unrivaled services in the analysis and processing of mass spectrometry data by leveraging our experience in computational science and our understanding of these powerful techniques. If you have any demands or questions, please don't hesitate to contact us.
References
For research use only, not intended for any clinical use.