Protein N-Terminal Sequencing
- Home
- Services
- Protein Sequencing
- Mass Spectrometry Based Protein Sequencing
- Protein N-Terminal Sequencing
Service Details
Protein N-terminal sequencing remains the method of choice for validating the N-terminal boundaries of recombinant proteins, identifying the N-terminal end of protease resistance structural domains, and identifying proteins isolated from species whose majority of genomes have not been sequenced. Meanwhile, ICH Q6B also requires N-terminal sequence confirmation for drugs. Compared with the Edman degradation method, in MS-based protein N-terminal sequencing, the N-terminal amino acids don't need to be repeatedly cleaved, and the proteins are directly pretreated by digestive enzymes into small fragments to analyze by mass spectrometry. Different peptides can generate fragmentions with different m/z and different spectra to obtain more N-terminal sequence information due to the different masses of most amino acid residues. In addition, mass spectrometry N-terminal sequencing is not affected by the N-terminal end of the protein, and which can also sequence N-terminal blocked and chemically modified amino acids.
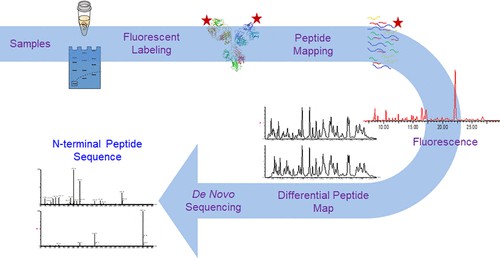 Fig. 1. Identification and sequencing of N-terminal peptides in proteins by LC-fluorescence-MS/MS (Malgorzata Monika Vecchi, et al., 2019)
Fig. 1. Identification and sequencing of N-terminal peptides in proteins by LC-fluorescence-MS/MS (Malgorzata Monika Vecchi, et al., 2019)
In the domain of proteomics, the N-terminus of a protein significantly influences its synthesis, structural configuration, and functional attributes. Utilizing the advanced capabilities of our mass spectrometry infrastructure, the team at Creative Proteomics has innovated a specialized approach for the precise sequencing of the N-terminal region of proteins. This methodology is adept at overcoming challenges associated with the analysis of proteins that exhibit N-terminal modifications or blockages, facilitating an in-depth characterization of proteins in their native or modified states.
We provide mass spectrometry-based protein N-terminal sequencing, which avoids the interference of N-terminus closure or modification sites and can achieve 100% coverage of the measured target protein sequence. For protein N-terminal sequencing, we developed a customized sequencing workflow.
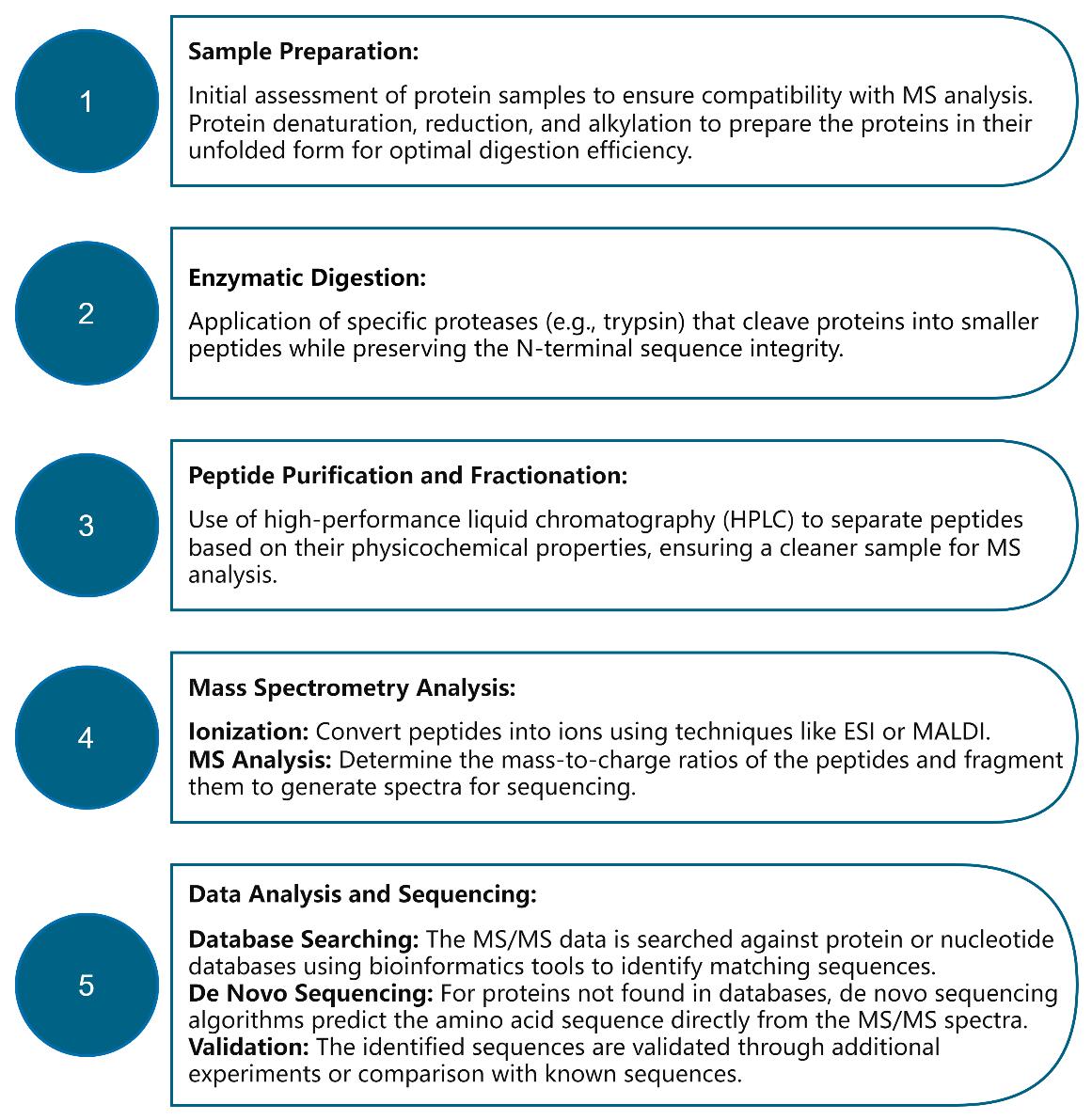 Workflow for N-Terminal Sequencing by Mass Spectrometry
Workflow for N-Terminal Sequencing by Mass Spectrometry
We also offer Edman degradation N-terminal sequencing services. Our Edman degradation service employs the classical technique of sequentially removing and identifying amino acids from the N-terminal end of peptides and proteins. This method is highly precise and remains the gold standard for N-terminal sequencing, especially suitable for:
 Fig. 2. Imaged N-terminal sequencing workflow.
Fig. 2. Imaged N-terminal sequencing workflow.
![]() The peptide coverage is as high as 100%, and up to 70 amino acid sequences at the N-terminal of the protein can be determined.
The peptide coverage is as high as 100%, and up to 70 amino acid sequences at the N-terminal of the protein can be determined.
![]() Automatic sequence analysis is more accurate.
Automatic sequence analysis is more accurate.
only need 1-10 μg protein sample can be detected with high sensitivity.
![]() Sequencing is not affected by N-terminal modifications such as N-terminal blocking, PEG and glycosylation.
Sequencing is not affected by N-terminal modifications such as N-terminal blocking, PEG and glycosylation.
Understanding the dynamics and degradation rates of MeCP2 protein variants in living cells is crucial for elucidating their roles in cellular processes and disease pathology. MeCP2 plays a vital role in transcriptional regulation and chromatin organization, and mutations in this protein are associated with neurodevelopmental disorders such as Rett syndrome.
The study utilized HEK293T and C2C12 cell lines transfected with wild-type (WT) and Ala2Val mutant MeCP2 constructs. Affinity-purified MeCP2 proteins were analyzed using gel-free purification and mass spectrometry techniques.
Cloning, Mutagenesis, Cell Culture, and Transfections:
Cell Harvesting and Protein Extraction:
Gel-Free Affinity Purification/Immunoprecipitation and Trypsin Digestion for Mass Spectrometry:
Mass Spectrometry to Determine MeCP2 PTMs:
Tryptic peptides were analyzed using a Q-Exactive™ Hybrid Quadrupole-Orbitrap™ mass spectrometer.
Cell Fixation and Confocal Imaging:
Fixed cells were stained with DAPI and imaged using confocal microscopy to study protein localization.
Fluorescence Recovery After Photobleaching (FRAP):
Cell Viability Test:
SK-N-SH cells were treated with cycloheximide (CHX) and imaged to assess cell viability.
Cycloheximide Chase Assay:
Neuroblastoma cells were transfected with MeCP2 constructs and treated with CHX for western blot analysis.
Real-Time Bleach-Chase Assays:
HEK293T cells were transfected with MeCP2 constructs and subjected to bleach-chase experiments to study protein degradation rates.
Data Analysis:
Various software tools were employed for data analysis, including Olympus FluoView, MATLAB-based easyFRAP, Microsoft Excel, and GraphPadTM.
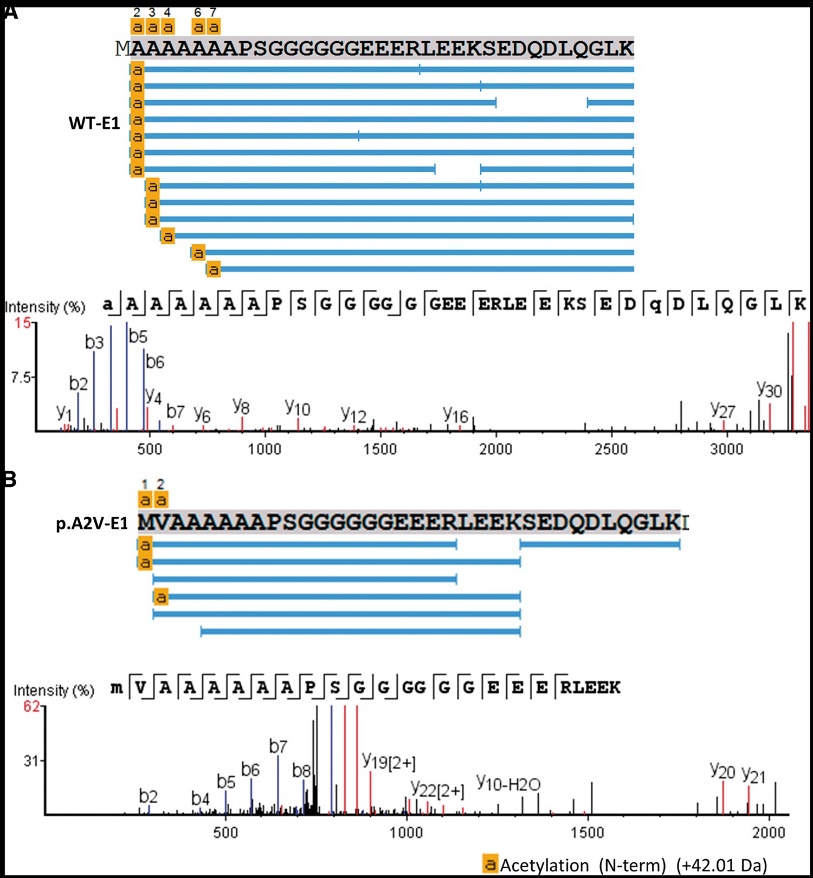 Mass spectrometry sequencing of the N-terminal of the MeCP2 protein (in vitro).
Mass spectrometry sequencing of the N-terminal of the MeCP2 protein (in vitro).
The study revealed differences in the mobility, degradation rates, and half-life of WT and Ala2Val mutant MeCP2 proteins in living cells. The Ala2Val mutation led to slower recovery rates and higher degradation rates compared to WT MeCP2, suggesting potential implications for disease pathology. Additionally, no significant differences were observed in protein-chromocenter localization or overall chromatin organization between WT and mutant MeCP2 constructs. These findings provide insights into the functional consequences of MeCP2 mutations and their relevance to neurodevelopmental disorders.
Reference
For research use only, not intended for any clinical use.