Top-Down Based PTMs Characterization Services
- Home
- Applications
- Proteomics Analysis Services
- Protein Post-Translational Modification Analysis Services
- Top-Down Based PTMs Characterization Services
Service Details
Top-down is a analysis method that sequencing or characterizing intact proteins without any truncation, which the method allows analysis of intact proteins without the need to digest peptides.
Top-down mass spectrometry has unique advantages in identifying protein forms with multiple post-translational modifications and/or unknown alterations. The process that intact proteins are introduced into the mass spectrometer for analysis reduces the complexity of the initial sample and crucially preserves information about combinatorial PTMs, sequence variants, alternative splicing and so on. Thus, this strategy can preserve labile modification motifs that are largely lost when using shotgun and CID for fragmentation in protein therapeutic assays, such as monoclonal antibodies and recombinant proteins.
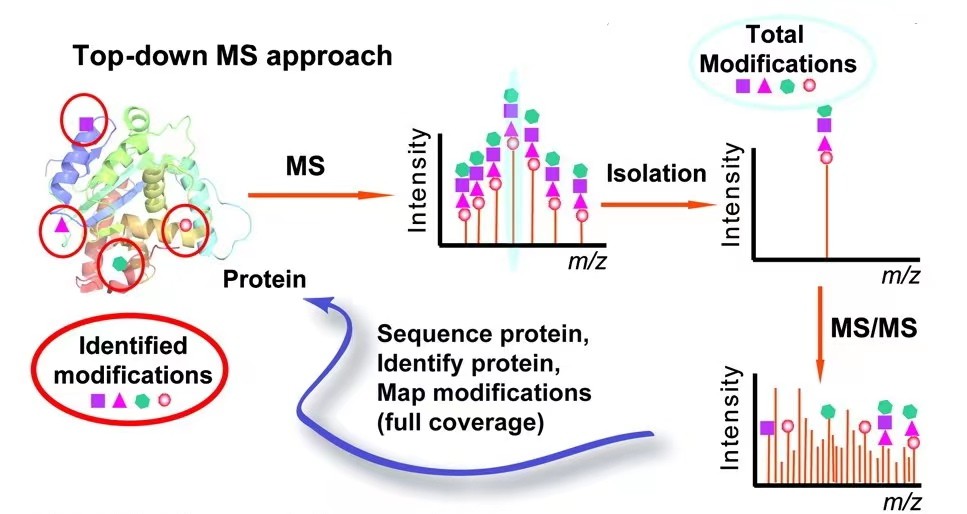 Fig. 1. Top-down for
protein PTM characterization. (Zhang H, et al., 2011)
Fig. 1. Top-down for
protein PTM characterization. (Zhang H, et al., 2011)
With advanced mass spectrometry platforms and rich experience in proteomics analysis, Creative Proteomics provides you with top-down mass spectrometry analysis services for protein post-translational modification to help your research projects.
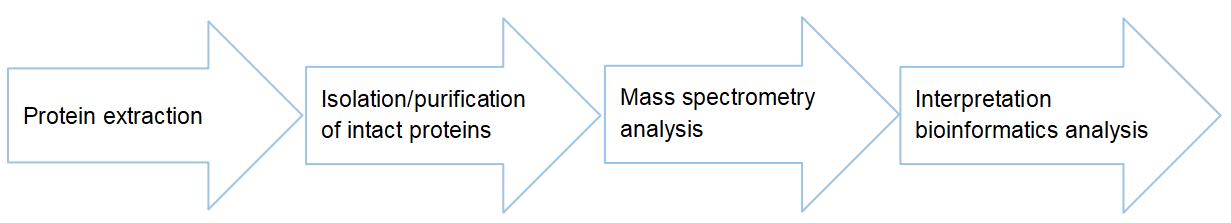
At Creative Proteomics, our scientists and technicians with experienced proteomics take a top-down approach to decode your protein, while preserving full PTMs information. Throughout the service cycle, intact proteins are ionized by electrospray ionization (ESI) or laser matrix desorption ionization (MALDI). The resulting ions are fragmented by collision-induced dissociation (CID), electron capture dissociation (ECD), or electron transfer dissociation (ETD) and analyzed in a tandem mass spectrometer to obtain protein information.
Notably, top-down proteomics stills under development and confronts with technical challenges in sample preparation, but the unique advantages mentioned above still make it a promising tech solution for characterization of post-translational modifications. It is complementary to bottom-up analysis and enables large-scale analysis of protein post-translational modifications, thus providing comprehensive and high-quality services for your PTMs analysis.
With cutting-edge equipment and expertise in protein sequencing and mass spectrometry, we provide one-stop top-down based PTMs characterization services. You only need to tell us the purpose of your experiment and send your samples to us, we will take care of all the follow-up matters of the project.
Thanks to our powerful mass spectrometry sequencing platform, Creative Proteomics provides top-down based PTMs characterization services for global researchers. Please feel free to contact us with any questions regarding protein post-translational modification analysis.
References
For research use only, not intended for any clinical use.