Peptide Mapping
- Home
- Services
- Protein Sequencing
- Mass Spectrometry Based Protein Sequencing
- Peptide Mapping
Service Details
Peptide mapping is an analytical technique widely used in the biopharmaceutical industry throughout the development of therapeutic proteins, primarily for the identification and verification of protein primary structure (amino acid sequence and chemical modifications). For recombinant protein drugs, peptide mapping analysis can be used to identify and characterize primary structures. In addition, the drug discovery process, various post-translational modifications (PTMs) produced by antibodies, degradation of protein molecules before storage, and contamination of protein samples, etc., directly affect the efficacy, stability and safety of drugs. Peptide map analysis is also the manufacture of these Process monitoring and Quality Assurance/Quality Control (QA/QC) are important techniques to ensure that there are no undesired changes in the product. Peptide mapping identifies proteins by first determining the molecular weights of the peptides and then matching those peptide masses to the protein's calculated theoretical mass.
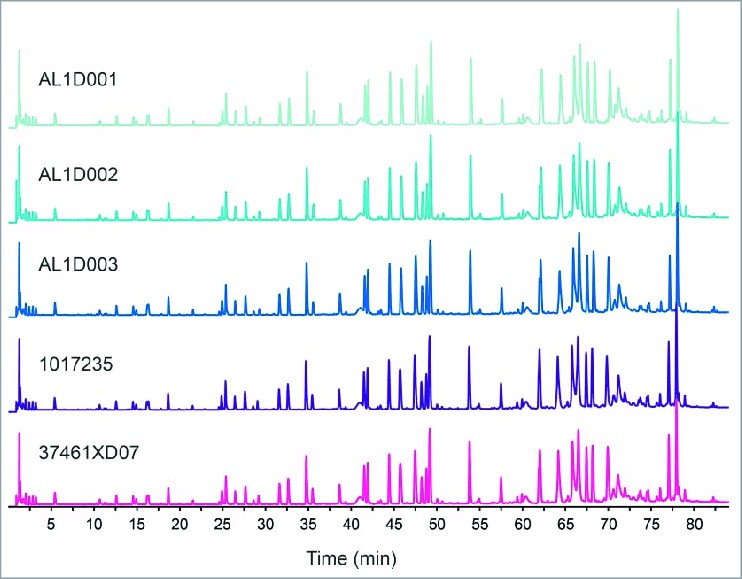 Fig. 1. Comparison of total UPLC peptide
mapping profiles. (Laurent M, et al., 2017)
Fig. 1. Comparison of total UPLC peptide
mapping profiles. (Laurent M, et al., 2017)
Creative Proteomics has developed a liquid chromatography and tandem mass spectrometry (LC-MS/MS)-based peptide mapping platform for protein primary structure confirmation. The peptide mapping analysis platform we offer uses tandem mass spectrometry instruments with extremely high resolution, combined with dedicated bioinformatics software that allows real-time assessment of coverage, to provide you with the most professional, rapid detection service. And we can provide peptide mapping services to analyze the sequence of biopharmaceuticals and confirm the complete expression of recombinant proteins in compliance with ICH Q6B. Because the peptide mapping service compares peptide masses to protein databases, only proteins in the database can be identified using this method. Proteins whose sequences cannot be found in protein databases, such as antibodies or novel proteins, can be inferred by our de novo sequencing service.
At Creative Proteomics, we have developed a complete workflow for protein peptide mapping analysis.
 Fig. 2. Workflow of the de novo peptide sequencing.
Fig. 2. Workflow of the de novo peptide sequencing.
Ⅰ. Amino Acid Sequence Analysis
Peptide mapping is typically performed on isolated proteins or protein mixtures, and identification of proteins using peptide mapping requires digestion of proteins into peptides prior to MS analysis. The first choice for many proteomics workflows is to generate peptides with trypsin. However, for certain regions of the protein, trypsinization may produce peptides that are too small or too large, depending on the situation, to be correctly detected by the machine. This would result in a lack of sequence information for these regions. To prevent this, our experts first analyze the amino acid sequence of your protein, selecting trypsin or other enzymes (Lys-C, Glu-C, etc.) to ensure 100% sequence coverage of the peptides.
Ⅱ .Multiple Enzyme Digestion and LC-MS/MS
Once the choice of proteases is set, we will perform the real-life digestion of your protein in the lab. Peptides resulting from each digestion will be analyzed on a short LC-MS/MS gradient using a data-dependent acquisition workflow.
Ⅲ. Reversr Sequence Conformation by Massanalyse
The molecular weight of the peptide is obtained after digestion.Then, The experimental mass of these peptides was compared to the mass produced by computer digestion of the protein or translated nucleic acid sequences contained in the database, resulting in a list of regions covered by the peptides in each protease. The experimental data is matched multiple times to the database one mass, and finally, we will pool and align each confirmed peptide from each digest on the protein's theoretical sequence to obtain the final peptide map.
Ⅳ. Data Report
The peptide map data report includes a list of each detected peptide that separated by protease. We will provide total protein coverage, along with the confidence associated with each peptide.
Peptide mapping can be used to document a range of properties of a target protein. At Creative Proteomics, our clients widely use our peptide mapping services in the following areas, including but not limited to.
At Creative Proteomics, our professional and experienced scientists not only provide you with high-quality peptide mapping services to determine the exact amino acid sequence of your target protein, but provide a comprehensive set of information to help the drug development process. Please contact us to discuss your specific needs if you have any questions about project.
References
For research use only, not intended for any clinical use.