Protein De Novo Sequencing and Mutation Analysis
- Home
- Services
- Protein Sequencing
- Mass Spectrometry Based Protein Sequencing
- Protein De Novo Sequencing and Mutation Analysis
Service Details
Protein molecules play a key role in kit development, diagnostic reagent development, and drug discovery. Traditional protein sequence analysis technology, whether based on MALDI-TOF mass spectrometer or nanoLC-MS/MS platform, requires the help of a sequence database including the identified protein in the process of protein identification, and then the molecular weight data determined by mass spectrometry is compared with. The molecular weight data obtained after the theoretical sequence fragmentation of the database is compared to realize the sequencing and identification of the protein. However, in the actual protein sequence analysis process, there are many protein information that are not included in the existing database, such as a completely new protein that has not been reported, or a protein purified from a completely new species, or partially mutated. For protein drugs, the protein sequence of which cannot be completed only by database search.
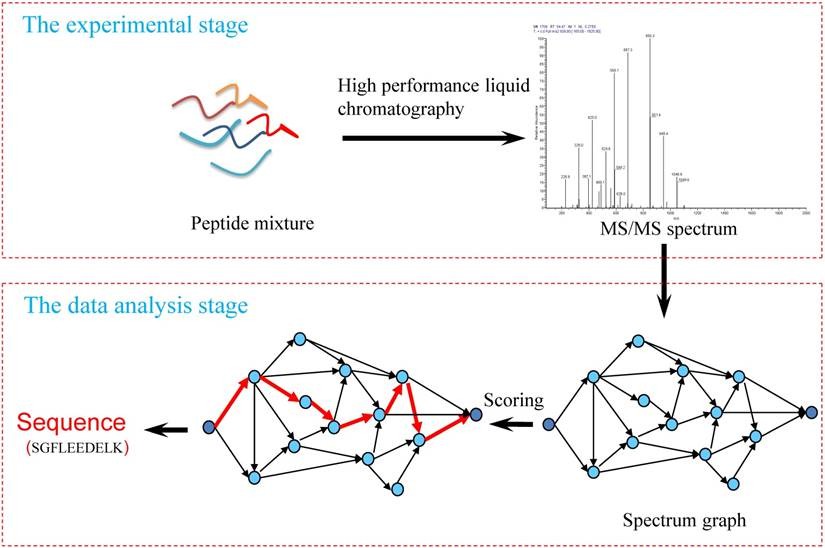 Fig. 1. Workflow of the de novo peptide sequencing. (Chuang Li, et al., 2019)
Fig. 1. Workflow of the de novo peptide sequencing. (Chuang Li, et al., 2019)
Creative Proteomics has established a new generation of protein de novo sequencing and mutation analysis platform to achieve fast and accurate high-quality sequence analysis of protein primary structure on the basis of the company's advanced mass spectrometry platform and rich experience in bioinformatics analysis. In the process of protein sequence determination, different proteases were selected for digestion to achieve100% full sequence splicing of protein molecules by using the complementarity between different digestion peptides.
Here, we provide protein de novo sequencing and mutation analysis services as follows:
Our established mass spectrometry sequencing platform has been widely chosen by our cusyomer for the following unique features.
Our protein de novo sequencing and mutation analysis platform is also well-suited for the following sequencing services.
At Creative Proteomics, we provide professional de novo protein sequencing and mutation analysis protein for global customers which was used for sequence and mutational analysis of unknown proteins. Our scientists can also design customized services. Please feel free to contact us If you are interested in our services, we look forward to with your cooperation.
References
For research use only, not intended for any clinical use.