- Services
- FAQ
- Case Study
- Related Services
- Support Documents
- Inquiry
What is Edman Degradation?
N-terminal sequencing by Edman degradation remains a valuable method for protein analysis with unique advantages. This cyclic process labels and cleaves one N-terminal amino acid at a time, identified via chromatography. The protein's N-terminal amino group reacts with phenyl isothiocyanate, forming a phenylthiocarbamoyl derivative. Under mild acidic conditions, a PTH-amino acid is released for identification. The released PTH amino acid is identified by high performance liquid chromatography (HPLC) or HPLC-MS. The remainder of the peptide is intact and available for another round of labeling and release.
Creative Proteomics provides precise N-terminal sequencing using the classical Edman degradation method, a cornerstone in protein analysis for accurately identifying N-terminal amino acids.
Technology Platform of N-terminal Sequencing by Edman Degradation Service
①Labeling the N-terminal amino acid: The N-terminal amino acid reacts with phenyl isothiocyanate (PITC), forming a phenylthiocarbamyl (PTC) derivative.
②Cleavage of the N-terminal amino acid: The labeled amino acid is cleaved under acidic conditions, exposing a new N-terminus for the next cycle.
③Identification of the cleaved amino acid: The cleaved amino acid is isolated and identified via high-performance liquid chromatography (HPLC). The PTC derivative is hydrolyzed into phenylthiohydantoin (PTH)-amino acids, which are then detected by UV absorbance or mass spectrometry based on their unique retention times.
④Iterative process: The steps of labeling, cleavage, and identification are repeated, allowing for the identification of one amino acid at a time from the N-terminus. This iterative process reconstructs the amino acid sequence by compiling the identified residues.
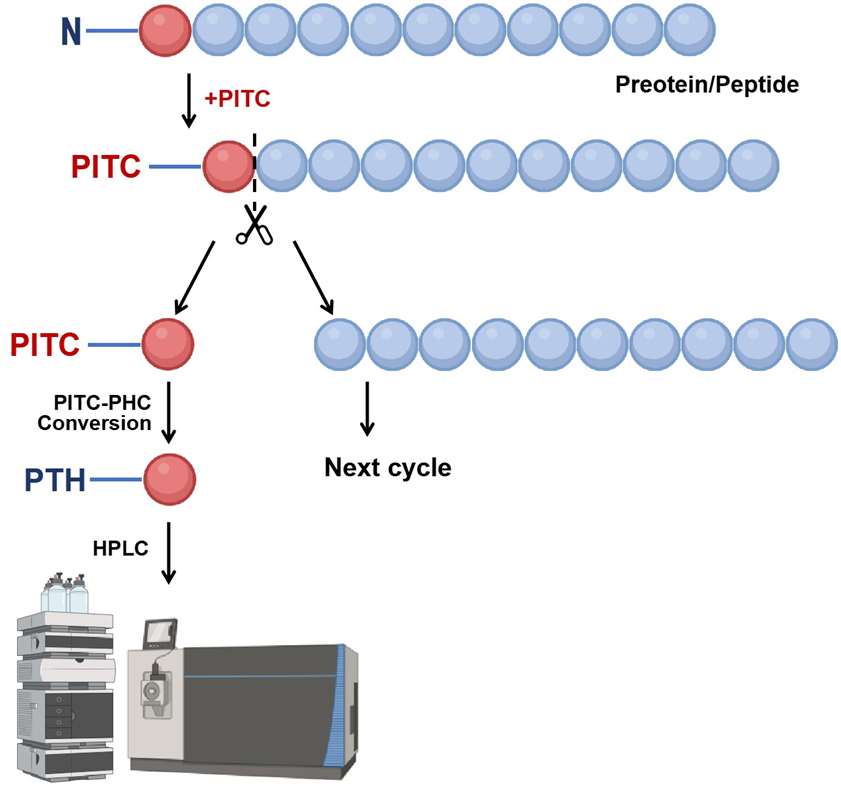 Figure 1. The workflow of the N-terminal sequencing via Edman degradation.
Figure 1. The workflow of the N-terminal sequencing via Edman degradation.Difference Between Edman Degradation and Sanger Sequencing
While Edman degradation is used to determine the N-terminal sequence of proteins, Sanger sequencing is a method for determining the sequence of nucleotides in DNA. The two techniques differ fundamentally in their substrates, mechanisms, and applications:
| Feature | Edman Degradation | Sanger Sequencing |
|---|---|---|
| Target Molecule | Proteins and peptides | DNA |
| Substrate | Free α-amino group at N-terminal | DNA template |
| Sequencing Mechanism | Chemical cleavage and HPLC | Chain termination and electrophoresis |
| Applications | Protein characterization, N-terminal sequence determination | Genetic analysis, DNA sequencing |
What Is the Characteristic of Edman Degradation?
Edman sequencing is not a sensitive method, requiring several picomoles and purified protein, to obtain a define sequence information. One important thing for Edman sequencing is that the α-amino group at the N-terminal end of the molecule should be unmodified before undergoing the cyclic sequencing process. Theoretically, it is possible to obtain sequence of the entire peptide or protein in a single sequencer run. However, multiple factors limit the amount of sequence in practice. With current technology, it is fairly routine to obtain at least 20 to 25 residues of sequence from the N-terminus of the proteins and peptides.
Applications of N-terminal Sequencing by Edman Degradation
- Verify the N-terminal boundary of recombinant proteins, particularly proteins larger than 40-80 kDa when highly accurate masses cannot be obtained by ESI-MS.
- Identifying mutations, truncations, and modifications such as acetylation or methylation.
- Characterizing novel proteins without the need for a reference database.
- Complementing structural studies with sequence-level insights.
Sample Requirements for N-Terminal Sequencing by Edman Degradation
| Requirement | Details |
|---|---|
| Sample Quantity | - Recommended: ≥200 pmol for 15 cycles or longer sequencing runs - Minimum: 25 pmol for 5 cycles |
| Sample Type | - Lyophilized protein or peptide samples - Liquid protein/peptide samples (10–100 µL) in volatile solvents (e.g., Milli-Q water, acetonitrile) - Protein bands blotted on PVDF membrane (≤ 3 × 6 mm) |
| Sample Purity | - At least 75–80% purity - Free amino acids, primary amines (e.g., Tris, glycine), SDS, salt, and other contaminants should be minimal |
| Cysteine Modification | Cysteine residues must be alkylated with iodoacetamide or 4-vinylpyridine for detection |
FAQ
Q: What is the maximum length of the peptide or protein that can be sequenced using Edman degradation?
A: Edman degradation is effective for sequencing the N-terminal up to approximately 40 amino acids. However, the length of the sequence that can be determined depends on several factors, including the sample amount, sample quality, and the protein's sequence. If longer sequences are required, proteins can be fragmented into smaller peptides, and each fragment can be sequenced individually.
Q: Is Edman degradation suitable for sequencing proteins from complex mixtures or crude extracts?
A: Edman degradation is most effective when the protein or peptide sample is purified. Crude extracts or complex mixtures typically contain a variety of proteins and other contaminants that can interfere with the sequencing process. It is highly recommended that the protein of interest be purified before submission to ensure high-quality sequencing results. SDS-PAGE or other purification methods (such as affinity chromatography) are commonly used to isolate the target protein prior to Edman sequencing.
Q: Can you provide additional data, such as peptide fragmentation patterns or secondary structure information?
A: Edman degradation primarily provides N-terminal sequence data. However, if you are interested in additional information like peptide fragmentation or secondary structure details, we recommend using mass spectrometry-based techniques, which offer broader proteomic coverage. If desired, we can complement Edman degradation with other proteomics services, such as peptide mapping, to provide a more comprehensive analysis of your protein.
Q: How does Edman degradation differ from mass spectrometry for protein sequencing?
A: Edman degradation and mass spectrometry (MS) are both powerful techniques, but they have distinct advantages and limitations:
Edman degradation: Directly sequences the N-terminal amino acids and is ideal for analyzing proteins where the sequence information is critical. It does not require a protein database and can accurately determine sequences even for novel proteins or proteins with mutations. However, it has a lower throughput and is limited to analyzing the N-terminal portion of the protein.
MS: Provides broader coverage of the protein sequence by detecting peptides generated from protein digestion, making it suitable for identifying larger proteins or for identifying proteins when their sequences are not known. However, MS relies on comparison to protein databases and may struggle with resolving ambiguous sequences, especially in cases where amino acids have similar masses.
Case Study
Case: Trop‐2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis
Background
Trop-2 (trophoblast cell-surface antigen-2) is a transmembrane glycoprotein involved in tumor growth and metastasis. It is strongly upregulated in various cancers but not typically mutated in the TACSTD2 gene. Trop-2 undergoes proteolytic cleavage, which activates its signaling functions. The study investigates the role of ADAM10-mediated cleavage at the R87-T88 site in Trop-2's activation and its subsequent effects on tumorigenesis and metastasis. The research is motivated by a lack of understanding of the molecular mechanisms behind Trop-2's cancer-promoting activities and its potential as a therapeutic target.
Methods
Cell Lines and Mutagenesis: Wild-type (wt) Trop-2 and a cleavage-impaired mutant (A87-A88 Trop-2) were transfected into various cell lines, including murine MTE4-14 and KM12SM colon cancer cells.
Cleavage and Activation Analysis:
Western blotting, flow cytometry, and cell-surface biotinylation assays were employed to assess Trop-2 cleavage at the R87-T88 site.
N-terminal Edman degradation was performed to confirm the cleavage site and verify the generation of specific Trop-2 fragments. This sequencing method allowed precise identification of the proteolytic processing at the R87-T88 cleavage site.
Proliferation and Metastasis Assays: In vitro cell growth was measured using proliferation assays, and in vivo tumor and metastasis growth were evaluated in murine xenograft models.
Inhibition Studies: The role of ADAM10 and MMP9 in Trop-2 cleavage was examined using specific siRNAs and inhibitors (e.g., ADAM10 inhibitor GI254023X).
Patient Sample Analysis: Trop-2 cleavage and ADAM10 expression were analyzed in breast cancer samples and normal tissues using Western blotting, histopathology, and proteomic approaches.
Proteomic and Phosphoproteomic Analysis: Advanced proteomic methods were applied to identify downstream signaling pathways activated by cleaved Trop-2 fragments.
Results
Trop-2 cleavage at R87-T88 increases with cell density and occurs specifically in cancer cells, excluding normal tissues and some well-differentiated tumors. A87-A88 Trop-2 mutants exhibited reduced cleavage and impaired cell growth compared to wild-type (wt) Trop-2.
ADAM10 was identified as the primary protease mediating Trop-2 cleavage at R87-T88. Inhibition of ADAM10 suppressed Trop-2 cleavage and tumor cell growth, while MMP9 inhibition showed no effect.
Cleaved Trop-2 activated signaling pathways (e.g., Src, MEK1, RSK1/2) crucial for cell proliferation and survival. Trop-2 cleavage was essential for tumor growth and metastasis in vivo, as demonstrated by reduced tumor size and metastasis in A87-A88 Trop-2 mutant models.
Trop-2 cleavage was prevalent in breast cancer samples, correlating with elevated ADAM10 levels and cancer progression, highlighting its potential as a biomarker and therapeutic target.
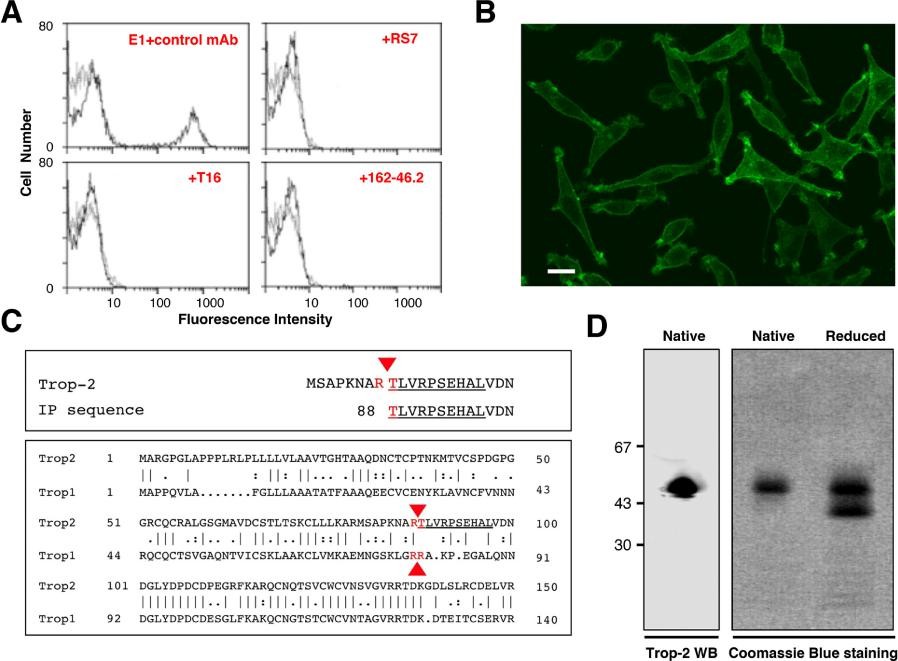 Figure 2. Purification, sequencing and analysis of Trop-2. (A) Reconstituted mixtures of 70% parental L cells and 30% Trop-2/L cells transfectants were used for competition studies of E1 with other anti-Trop-2 mAbs. (B) Reactivity of the E1 mAb with Trop-2. (C, top) Alignment of the amino-acid sequence obtained by Edman degradation (underlined) with the canonical Trop-2 sequence. (C, bottom) Alignment of the proteolytic sites of Trop-2 and Trop-1 [21] (red arrows). (D, left) Western blotting of Trop-2, as immunoprecipitated from ovarian cancer cells and purified by affinity chromatography over Sepharose–E1 mAb. (D, right) Coomassie blue staining of purified Trop-2 protein run in SDS-PAGE gradient gel under native or reducing conditions.
Figure 2. Purification, sequencing and analysis of Trop-2. (A) Reconstituted mixtures of 70% parental L cells and 30% Trop-2/L cells transfectants were used for competition studies of E1 with other anti-Trop-2 mAbs. (B) Reactivity of the E1 mAb with Trop-2. (C, top) Alignment of the amino-acid sequence obtained by Edman degradation (underlined) with the canonical Trop-2 sequence. (C, bottom) Alignment of the proteolytic sites of Trop-2 and Trop-1 [21] (red arrows). (D, left) Western blotting of Trop-2, as immunoprecipitated from ovarian cancer cells and purified by affinity chromatography over Sepharose–E1 mAb. (D, right) Coomassie blue staining of purified Trop-2 protein run in SDS-PAGE gradient gel under native or reducing conditions.References
- Gooley A A, et al. A role for Edman degradation in proteome studies. Electrophoresis, 1997, 18(7): 1068-1072. DOI: 10.1002/elps.1150180707
- Zhang G, et al. Overview of peptide and protein analysis by mass spectrometry. Current protocols in protein science, 2010, 62(1): 16.1. 1-16.1. 30. DOI: 10.1002/0471140864.ps1601s62
- Wilkins M R, et al. Rapid protein identification using N-terminal "sequence tag" and amino acid analysis. Biochemical and biophysical research communications, 1996, 221(3): 609-613. DOI: 10.1006/bbrc.1996.0643
- Trerotola M, Guerra E, Ali Z, et al. Trop‐2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis. Neoplasia, 2021, 23(4): 415-428. DOI: 10.1016/j.neo.2021.03.006
Related Services
Support Documents