- Services
- FAQ
- Demo
- Case Study
- Related Services
- Support Documents
- Inquiry
Understanding glycosylation is essential for biopharmaceutical innovation. At Creative Proteomics, our glycopeptide analysis services offer critical insights into glycan structures, glycosylation sites, and overall glycoprotein integrity—factors that directly affect drug safety, efficacy, and regulatory approval.
Using state-of-the-art instrumentation and validated workflows, we help clients uncover:
- Precise glycopeptide sequences
- Glycosylation types and site occupancy
- Structural details and relative abundance of glycans
This analysis supports key quality attributes required under ICH Q6B guidelines for glycoprotein characterization.
Why Choose Creative Proteomics?
We're more than a service provider—we're a strategic partner in your biologics development pipeline. Our team delivers:
- Fully traceable, GLP/cGMP-compliant data
- High-throughput, high-resolution mass spectrometry analysis
- End-to-end support from sample preparation to data interpretation
Our glycopeptide analysis service ensures compliance while accelerating decision-making during cell line development, biosimilarity assessments, and formulation optimization.
Our Glycopeptide Analysis Solutions Include:
- Sequencing of glycopeptides for structure elucidation
- Molecular weight characterization of glycosylated peptides
- Composition analysis of glycan types (e.g., high-mannose, hybrid, complex)
- Determination of glycosylation site occupancy and distribution
- Linkage-specific profiling, including terminal sialic acid configuration
Whether you're optimizing monoclonal antibody production or validating biosimilar comparability, our glycopeptide profiling services can help you navigate regulatory complexity and improve biologic performance.
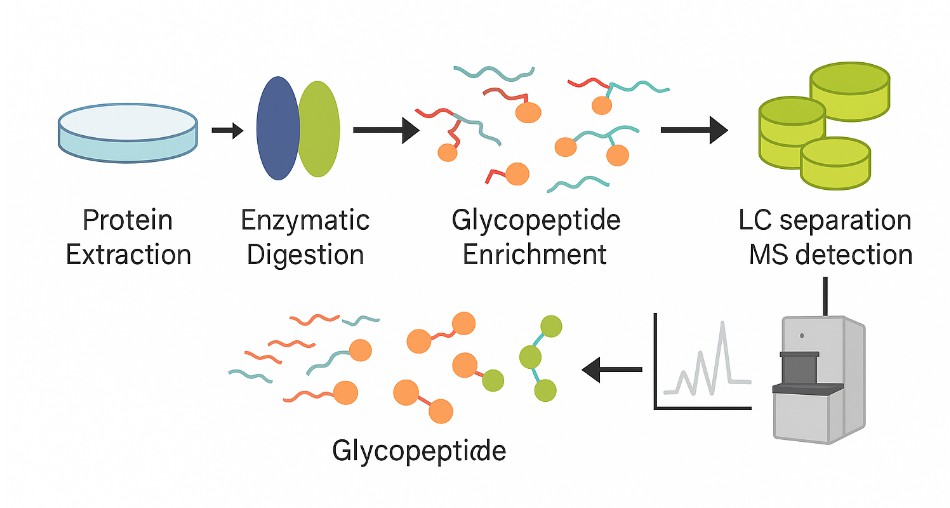
Workflow of Glycopeptide Characterization Using Mass Spectrometry
Sample Quality Control and Preparation
Upon receiving your samples, we perform concentration checks and buffer exchange (if necessary) to ensure compatibility with downstream mass spectrometry.
Enzymatic Digestion
Proteins are digested with trypsin or other appropriate enzymes to release glycopeptides while preserving glycosylation integrity.
Glycopeptide Enrichment
We apply targeted enrichment methods such as lectin affinity, HILIC, or PGC to selectively isolate both N-linked and O-linked glycopeptides.
Mass Spectrometry (LC-MS/MS) Analysis
High-resolution LC-MS/MS with advanced fragmentation (CID, HCD, or ETD) is used to detect glycopeptide sequences and locate glycosylation sites.
Data Processing and Structure Annotation
Spectra are interpreted using industry-leading software including Byonic, Byologic, SimGlycan, and GlypID 2.0 to annotate peptide sequences and glycan compositions.
Comprehensive Reporting
All results are compiled into a structured technical report, including glycopeptide maps, annotated spectra, and identification tables. Optional raw data and expert consultations are available upon request.
Technology Platform for Glycopeptide Analysis
At Creative Proteomics, we apply a multi-technique approach to deliver high-resolution insights into glycopeptide structures and glycosylation patterns. Our platform integrates sialic acid chemistry, HPLC, HILIC, lectin affinity separation, porous graphitic carbon (PGC) chromatography, collision-induced dissociation (CID), and advanced mass spectrometry (MS)—all optimised for accurate glycopeptide characterization.
1. Solid-Phase Chemoenzymatic Method (α2,3/α2,6 Sialic Acid Profiling)
This approach is ideal for resolving complex sialylation patterns:
- Glycoproteins are immobilized onto a solid-phase matrix.
- Selective derivatization of α2,3- and α2,6-linked sialic acids is achieved using an EDA-functionalized column.
- Derivatized products are purified via high-performance liquid chromatography (HPLC).
- Hydrophilic interaction chromatography (HILIC) is used for enrichment.
- On-bead digestion yields glycopeptides and peptides for LC-MS/MS.
- Spectral data are processed with Byonic and Byologic to map glycopeptide structures comprehensively.
2. Enzymatic Digestion + Lectin Affinity + PGC-CID-MS/MS
A versatile and widely used method for glycopeptide mapping:
- Glycoproteins are enzymatically digested using trypsin.
- Lectin affinity chromatography isolates glycopeptides with specific glycan motifs.
- Porous graphite carbon (PGC) enables efficient separation of glycans and peptides.
- Collision-induced dissociation (CID) is applied to fragment glycopeptides in the mass spectrometer.
- Data interpretation is automated using advanced software such as SimGlycan and GlypID 2.0, enabling accurate annotation of glycan-peptide structures.
This strategy is particularly suited for high-throughput glycosylation profiling and biosimilarity assessments, providing reliable site-specific glycosylation data.
With flexible platform configurations and expert method development, Creative Proteomics supports customizable glycopeptide analysis tailored to monoclonal antibody production, vaccine development, and other biologics programs.
Sample Requirements for Glycopeptide Analysis
| Category | Details |
|---|---|
| Acceptable Sample Types | - Purified glycoproteins (≥90% purity) - Cell lysates or culture supernatants - Biologics (e.g., mAbs, fusion proteins, viral glycoproteins) |
| Minimum Sample Amount | ≥100 µg total protein per sample Lower quantities accepted upon consultation |
| Storage Requirements | - Store at -80°C prior to shipment - Avoid freeze-thaw cycles |
| Shipping Instructions | - Ship on dry ice in leak-proof containers - Label samples clearly and securely |
| Required Information | - Sample ID and description - Protein concentration and buffer components - Analysis objectives (e.g., site-specific profiling, glycan mapping) |
Deliverables You Will Receive:
- A full experimental report detailing methods, results, and interpretations
- Annotated glycopeptide MS/MS spectra
- Peptide maps with glycosylation site assignments
- Tables of identified glycopeptides and associated glycan structures
- Optional relative quantification or glycoform distribution analysis
- Raw LC-MS data files (upon request)
Applications in Biopharmaceutical Development
Where Glycopeptide Analysis Makes a Difference
Our glycopeptide characterization service supports a wide range of biopharma applications, from early-stage candidate screening to regulatory submissions. Whether you are developing innovative biologics or assessing biosimilarity, our analysis provides the molecular detail needed to make confident decisions.
Common Use Cases:
Monoclonal Antibody Development
Verify Fc glycosylation patterns that impact antibody-dependent cell cytotoxicity (ADCC) and pharmacokinetics.
Biosimilarity Assessment
Compare site-specific glycosylation between reference and biosimilar products for regulatory compliance.
Vaccine Glycoprotein Characterization
Profile glycan structures on viral proteins to inform immunogenicity and stability studies.
Glycosylation Site Mutation Studies
Confirm changes in glycosylation occupancy and structure after site-directed mutagenesis.
Batch Consistency & Process Monitoring
Evaluate glycan heterogeneity across production lots as a key quality attribute under ICH Q6B.
Sialylation Linkage Profiling (α2,3 vs α2,6)
Distinguish between terminal sialic acid linkages, which may affect clearance rates and receptor binding.
Detection of Host Cell-Derived Glycoproteins
Identify and monitor glycoprotein impurities from CHO or other expression systems.
Enzyme Therapy Development
Analyze glycosylation of therapeutic enzymes to optimize activity, targeting, and half-life.
Frequently Asked Questions
Q: Can your service analyze both N-linked and O-linked glycopeptides?
A: Yes. We offer validated enrichment strategies that target both N-glycosylation and O-glycosylation. Clients may request combined analysis or specify the glycopeptide type of interest.
Q: Is it possible to determine glycosylation site occupancy?
A: Absolutely. Our mass spectrometry-based workflow provides site-specific resolution, allowing for the detection of both occupied and unoccupied glycosylation sites.
Q: Can you differentiate between α2,3 and α2,6 sialic acid linkages?
A: Yes. We apply chemoenzymatic labeling and specialized LC-MS techniques to distinguish between these two common sialylation types.
Q: What sample types are acceptable for analysis?
A: We can analyze a variety of samples, including purified glycoproteins, cell lysates, and formulation buffers. We recommend removing high concentrations of salts, detergents, or stabilizers prior to submission.
Q: Are the results suitable for regulatory submissions or publication?
A: Yes. Our workflows are performed under well-documented quality protocols, and deliverables include structured reports with annotated spectra, peptide maps, and identification tables suitable for regulatory or publication use.
Q: Do you offer support with data interpretation?
A: Yes. Our team includes experienced scientists who can assist with result interpretation, method selection, and project planning based on your research goals.
Q: Can I customize the analysis workflow?
A: Definitely. We offer flexible service models, allowing you to select specific glycan targets, glycopeptide types, or data formats. Custom method development is available upon request.
Q: What if I'm unsure about sample preparation?
A: No problem. We provide sample submission guidelines and can offer pre-processing services such as buffer exchange, desalting, or protein quantification to ensure compatibility.
Demo
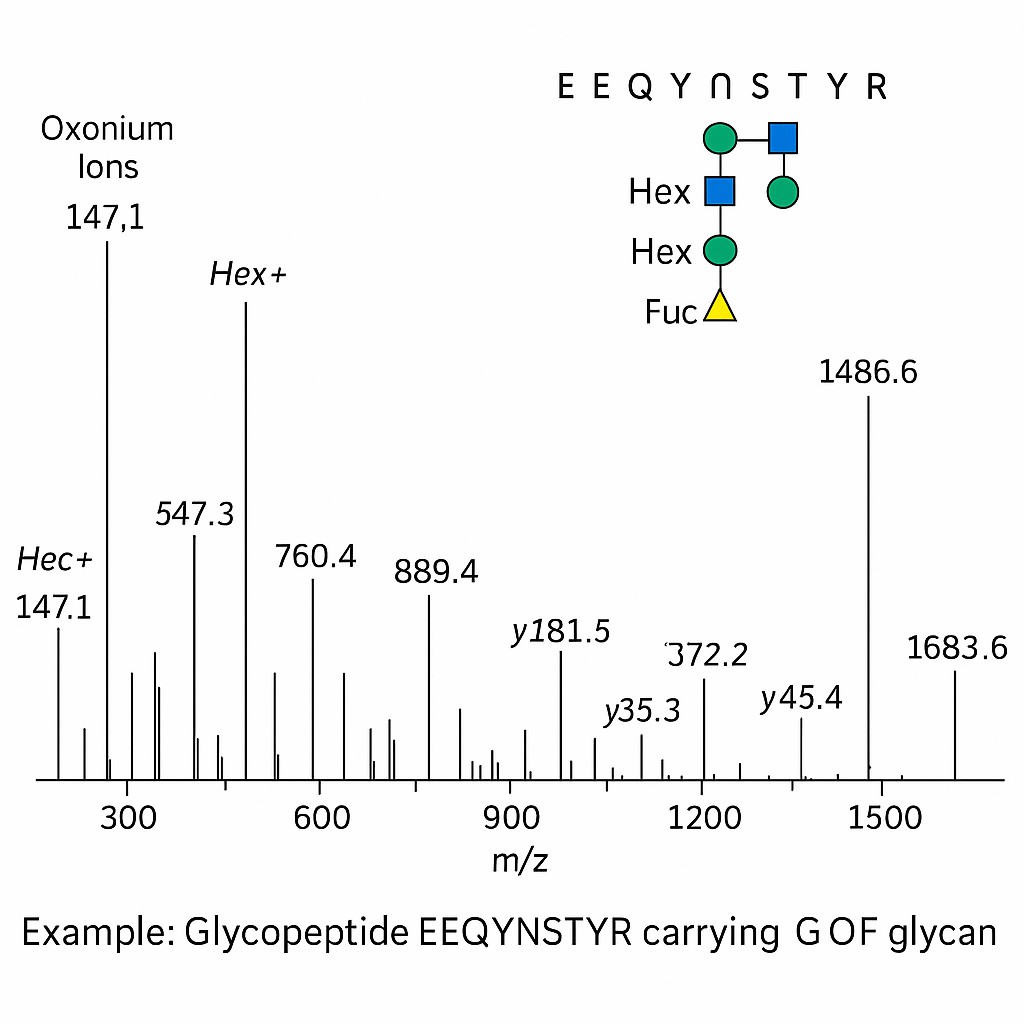
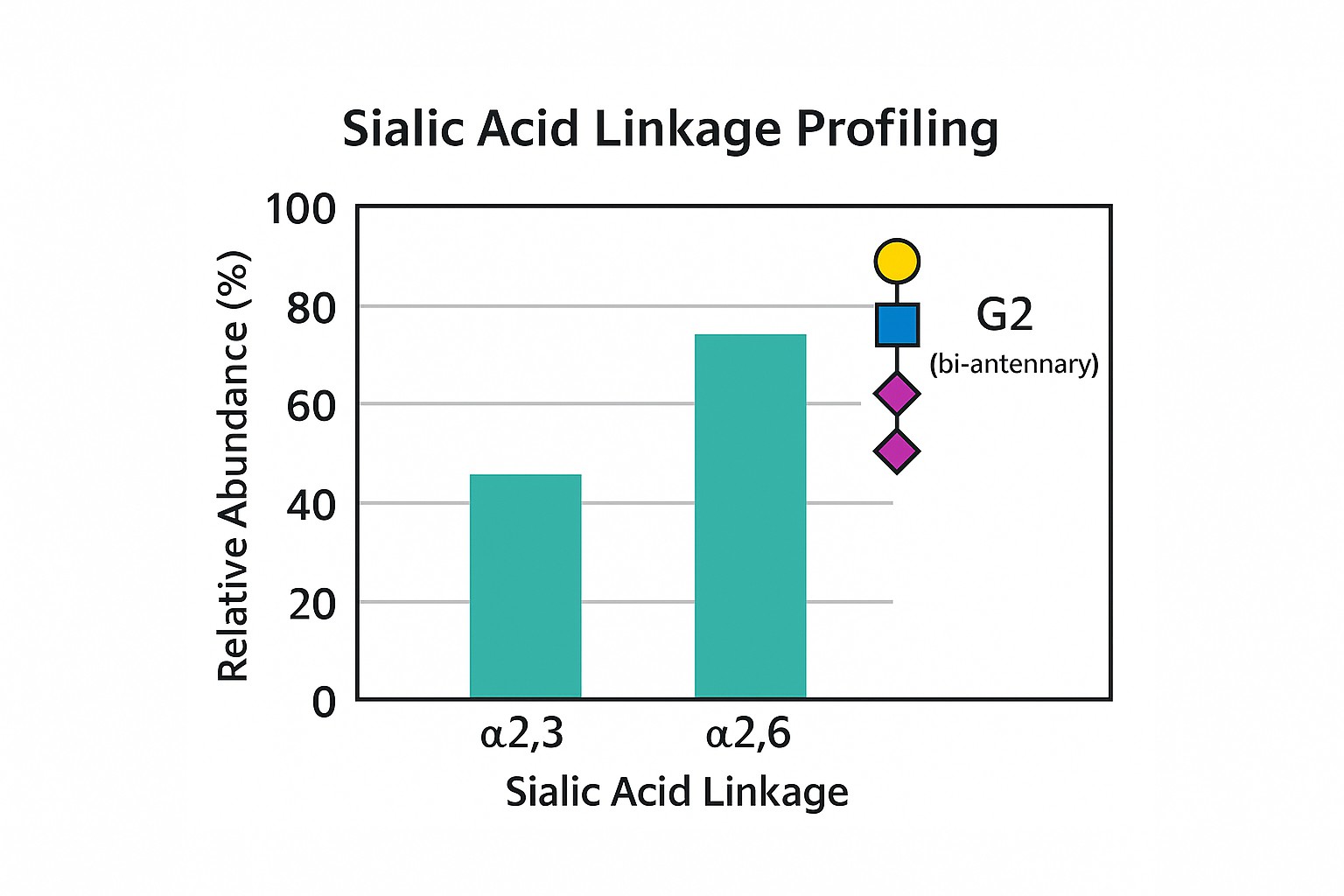
Case Study: Site-Specific Glycosylation Profiling in Prostate Cancer
doi: 10.1007/s13361-018-1931-0
Project Overview
This study aimed to perform LC-MS/MS-based intact N-glycopeptide analysis on human prostate cancer tissues and their adjacent non-cancerous counterparts to uncover site-specific heterogeneity in protein glycosylation associated with prostate cancer.
Analysis Approach
Utilized liquid chromatography-tandem mass spectrometry (LC-MS/MS) to analyze clinical prostate cancer tissues and matched non-cancerous tissues.
Employed Byonic software for glycopeptide identification and quantification.
Compared glycosylation sites between cancerous and non-cancerous tissues to identify cancer-associated glycosylation changes.
Key Findings
Multiple N-glycosylation sites exhibited distinct glycosylation patterns in prostate cancer tissues compared to non-cancerous tissues, suggesting a significant role of glycosylation in cancer progression.
Specific glycosylation sites demonstrated notable heterogeneity in cancer tissues, indicating their potential as biomarkers for prostate cancer diagnosis or therapeutic targets.
Certain glycan structures were enriched in cancer tissues, which may be linked to tumor invasiveness and metastatic potential.
This case study underscores the importance of site-specific glycosylation analysis in cancer research and demonstrates the capability of mass spectrometry techniques in revealing tumor-associated glycosylation alterations.
References
- Yang S, Wu W W, et al . Identification of Sialic Acid Linkages on Intact Glycopeptides via Differential Chemical Modification Using IntactGIG-HILIC. J Am Soc Mass Spectrom. 2018, 29(6):1273–1283. doi: 10.1007/s13361-018-1931-0
- Woodin C L, Maxon M, et al. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst. 2013, 138(10):2793–2803. doi: 10.1039/c2an36042j
- Cao L, Qu Y, et al. Intact glycopeptide characterization using mass spectrometry. Expert Rev Proteomics. 2016, 13(5):513–522. DOI: 10.1586/14789450.2016.1172965
Related Services
Support Documents