- Service
- Case Study
- FAQ
- Related Services
- Inquiry
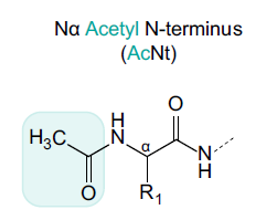
N-terminal acetylation—where an acetyl group is added to the N-terminal amino group of a protein—is one of the most prevalent post-translational modifications (PTMs) in biologics. Though small in structure, this modification can significantly alter a protein's behaviour. It may influence a molecule's charge, folding, hydrophobicity, stability, and even its interaction with other biomolecules.
In the context of drug development, understanding N-acetylation is critical. This modification can:
- Affect protein identity and function
- Impact pharmacokinetics and metabolism
- Contribute to polymorphic variability in patient response
More importantly, overlooking N-acetylation during characterization can compromise manufacturing consistency, product quality, and downstream safety.
At Creative Proteomics, we offer specialised N-acetylation analysis services designed for biopharmaceutical developers. In compliance with ICH Q6B guidelines, our services provide:
- Identification and quantification of N-terminal acetylation
- Detailed analysis of structural integrity and modification patterns
- Support for regulatory submissions using PharmEu and USP<1047> standards
Whether you're analysing a recombinant protein therapeutic or a next-generation biologic, our platform delivers high-resolution insights into N-terminal modifications—helping you ensure consistency, efficacy, and regulatory alignment throughout your development pipeline.
Comprehensive N-acetylation Analysis Services
At Creative Proteomics, we offer a full suite of N-acetylation analysis services tailored to the needs of pharmaceutical and biotech researchers. Whether you're developing biologics, studying metabolic pathways, or exploring protein-drug interactions, our services provide high-resolution insights into N-terminal acetylation events across both eukaryotic and prokaryotic systems.
Our Core N-Acetylation Services Include (but Are Not Limited To):
- N-terminal acetylation identification and profiling
- Degree of N-acetylation quantification
- Acetylation site mapping
- Quantitative analysis of acetylated peptides
- Drug metabolism studies involving acetylated compounds
- Drug–protein interaction analysis for acetylated molecules
- PK/PD studies of acetylated drugs
- Metabolomics profiling of acetylated compounds
Advanced Technology Platform
We use a robust, multi-step analytical workflow to ensure precision and reproducibility in every project:
Sample Preparation
- Proteins are extracted from lysed cells (eukaryotic or prokaryotic)
- Digestion is carried out using detergent-free trypsin
- Removal of N-pyroglutamate ensures cleaner peptide pools
Peptide Separation
- Peptides are fractionated using multi-dimensional chromatography
Enrichment of N-Acetylated Peptides
Depending on your project needs, we offer several enrichment options:
- COFRADIC (Combined Fractional Diagonal Chromatography)
- SCX (Strong Cation Exchange Chromatography)
- CNBr-activated Sepharose resin, a simpler method that remains efficient across varying digest-to-resin ratios
Mass Spectrometry Analysis
- Enriched peptides are analysed by high-resolution MS
- To enhance identification, we incorporate Singly Charged Ion Inclusion (SCII), enabling the detection of singly charged precursors often missed in conventional methods
This integrated platform, combining HPLC and MS, delivers sensitive, accurate, and reproducible data, making it ideal for in-depth structural and functional studies of acetylated proteins and peptides.
Workflow of Our Services

Why Choose Our N-acetylation Analysis Service?
Key Advantages
✅ High Sensitivity
Our method can detect N-terminal acetylation even in low molecular weight proteins, enabling more accurate profiling of subtle modifications.
✅ State-of-the-Art Equipment
We leverage industry-leading instruments to ensure exceptional resolution and reliability:
- SHIMADZU LC-20AT high-performance liquid chromatography
- Thermo Scientific™ Q Exactive™ Hybrid Quadrupole-Orbitrap™ mass spectrometer
✅ Optimized Analytical Platform
Using automated chromatography systems designed specifically for modification analysis, we reduce sample volume requirements while improving both efficiency and scalability.
✅ Fast Turnaround
Expect a full analytical report—including raw data, method details, and conclusions.
✅ Customised Protocols
Our team tailors buffer systems, enrichment methods, and MS settings based on your specific sample type and research goals.
Expert Support, Every Step of the Way
Our experienced analytical scientists deliver:
- Full confirmation of N-acetylation across known or novel protein targets
- Quantitative results on the degree of acetylation
- Clear, actionable reports with all protocols and raw data included
- Personalised support to help you troubleshoot and optimise your analytical strategy
Whether you're investigating post-translational modifications in biologics or exploring acetylation's impact on drug function, we're here to provide rapid, reliable, and research-driven insights.
Sample Requirement
| Parameter | Requirements | Notes |
|---|---|---|
| Sample Type | Recombinant protein, cell lysate, purified biologics | Supports both prokaryotic and eukaryotic sources |
| Protein Concentration | ≥ 0.5 mg/mL (≥ 100 µg/sample) | Recommendation based on typical proteomics needs |
| Sample Volume | ≥ 50 µL | Ensures sufficient material for chromatographic separation and MS analysis |
📝 Notes:
Recombinant protein may come from various expression systems (E. coli, yeast, mammalian cells).
Cell lysate requires at least ~100 µg total protein per condition for reliable PTM detection.
Purified biologics in buffer are acceptable; mention buffer components (e.g., salts, detergent).
Volume ≥50 µL supports multiple injections and enrichment steps in our workflow.
Client Case Study: HDAC4's Role in DNA Damage Response & Senescence
Background
Cellular senescence—permanent cell-cycle arrest—is driven by DNA damage accumulation and epigenetic shifts. Our client investigated how HDAC4, in concert with HDAC1/2, influences homologous recombination (HR) and modulates acetylation of histone H2B at lysine 120 (H2BK120ac). They aimed to clarify how HDAC4 loss disrupts DNA repair, leading to senescence.
Technical Methods
Chromatin immunoprecipitation (ChIP-qPCR & ChIP-seq) to profile H2BK120ac, H2BK120ub, H4K16ac, and γH2AX at DNA breaks.
Co-immunoprecipitation (Co-IP) and in vitro deacetylation assays confirmed HDAC4, HDAC1, and HDAC2 complex formation and functional activity.
HR efficiency assays using I SceI–induced double-strand breaks in U2OS/TRI DR cells, comparing wild-type vs H2BK120Q (acetyl-mimetic) histones.
Senescence models: RAS-induced senescence in BJ hTERT cells ± enforced HDAC4 expression; γH2AX immunoblotting quantified DNA damage spread.
Results
HDAC4 forms a functional complex with HDAC1/2, deacetylating H2BK120. HDAC4 deficiency → ↑ H2BK120ac, impaired recruitment of BRCA1/CtIP to DNA lesions → defective HR repair.
Acetyl-mimetic mutation (H2BK120Q) mirrors HDAC4 loss, significantly reducing HR repair efficiency .
During oncogene-induced senescence, HDAC4 degradation is correlated with increased γH2AX, spread of DNA damage, and elevated H2BK120ac.
Re expression of HDAC4 in senescent cells limits the genomic spread of γH2AX, lowers H2BK120ac, and boosts HR at damage sites.
 HDAC4 influences the epigenetic landscape at the DSBs.
HDAC4 influences the epigenetic landscape at the DSBs.Conclusion
Our platform enabled high-resolution mapping of histone modifications and DNA repair dynamics. We provided:
- Targeted ChIP-seq and quantitative ChIP-qPCR for H2BK120ac/ub and γH2AX across the genome.
- Biochemical validation of enzymatic deacetylation via in vitro assays.
- Functional HR assays demonstrating the direct impact of H2BK120 acetylation on DNA repair capacity.
Impact: This study uncovered how the HDAC4–HDAC1/2 complex safeguards genomic integrity and prevents senescence—via targeted epigenetic regulation of H2BK120 acetylation and homology-directed repair.
Frequently Asked Questions (FAQ) – N-acetylation Analysis Service
Q: What types of samples are suitable for N-acetylation analysis?
A: We support a wide range of sample types, including recombinant proteins, cell lysates (from prokaryotic and eukaryotic sources), and purified biologics. Our platform can be tailored to handle complex matrices depending on your project goals.
Q: Can your service distinguish between N-terminal acetylation and lysine acetylation?
A: Yes. Our workflows are designed to selectively enrich and identify N-terminal acetylation events, differentiating them from lysine (internal) acetylation through targeted enrichment and high-resolution MS.
Q: How sensitive is your platform in detecting low-abundance acetylated proteins?
A: Our integrated HPLC-MS system is optimized for high sensitivity, allowing for the detection and quantification of low-abundance N-acetylated peptides, even in challenging sample backgrounds.
Q: Do you provide site-specific acetylation information?
A: Absolutely. We map N-acetylation sites at the peptide level, providing detailed localization and confidence scores to support functional interpretation.
Q: Is your N-acetylation analysis service compliant with regulatory standards?
A: Yes. Our methods align with international guidelines such as ICH Q6B, PharmEu, and USP<1047>, making them suitable for characterization studies in support of regulatory submissions.
Q: Can you assist in selecting the most appropriate enrichment method for my sample?
A: Certainly. Based on your protein type, buffer conditions, and experimental goals, our scientists will recommend the most suitable enrichment strategy—such as COFRADIC, SCX, or CNBr-activated resin.
Q: What information is included in the final analytical report?
A: Our reports include raw and processed MS data, identified acetylation sites, quantification results, enrichment protocols, and interpretation support—delivered in a clear, publication-ready format.
Q: Can the analysis support research on drug–protein interactions or PK/PD studies?
A: Yes. We offer specialized workflows to study acetylated drug-protein interactions and support PK/PD or metabolomics studies involving acetylated compounds.
Q: Do you offer customization for specific protein targets or workflows?
A: Yes, all services are customizable. From sample prep to data analysis, we tailor each project to your protein target, sample type, and downstream research objectives.
Q: How do I get started with submitting samples for analysis?
A: Simply contact our technical team with your project details. We'll guide you through sample preparation, shipping requirements, and all necessary documentation.
References
- Protein Acetylation Methods and Protocols (2013). Hake S.B., Janzen C.J. (Eds). Hatfield, Hertfordshire (UK): Humana Press.
- Rioux N., Mitchell L.H., Tiller P., et al., Structural and kinetic characterization of a novel N-acetylated aliphatic amine metabolite of the PRMT inhibitor, EPZ011652. Drug Metab Dispos, 43:936–943, July 2015. DOI: 10.1124/dmd.115.064014
- Dadou S.M., El-Barghouthi M.I., Alabdallah S.K., et al., Effect of Protonation State and N-Acetylation of Chitosan on Its Interaction with Xanthan Gum: A Molecular Dynamics Simulation Study. Marine Drugs, 2017, 15, 298. DOI: 10.3390/md15100298
- Ree R., Varland S., and Arnesen T., Spotlight on protein N-terminal acetylation. Experimental & Molecular Medicine, (2018) 50:90. DOI: 10.1038/s12276-018-0116-z
Related Services