- Services
- Demo
- FAQ
- Case Study
- Related Services
- Support Documents
- Inquiry
What is Surface Plasmon Resonance (SPR)?
Surface plasmon resonance (SPR) affinity assay is an optical-based label-free detection technique for real-time detection of molecular interactions between two or more molecules. It offers high throughput, flexibility, and sensitivity for drug discovery and screening. For example, it has powerful advantages in screening small molecules or peptides (enzymes or antibodies, etc.), enabling rapid and accurate screening of candidate drug molecules to save costs for subsequent studies.
Based on the principle of SPR, Creative Proteomics provides accurate and rapid affinity assays. The throughput, flexibility and sensitivity of SPR affinity assays make it possible for researchers to study any binding, including: antigen-antibody affinity assays, protein-small molecule and peptide affinity assays, protein-small molecule drug affinity assays, etc. It is possible to study a wide range of molecules, from ions, fragments and small molecules to proteins and viruses.
How does SPR Work?
SPR affinity detection detects molecular interactions by tracking signal changes on a sensor chip. the SPR signal is measured in reaction units (RU), which is approximately equivalent to a protein surface concentration of 1 pg/mm2. During the interaction of two molecules, the SPR signal generates a sensing map, which can be visualized in real time (Figure 1). The general binding reaction consists of three phases: binding, equilibrium and dissociation. Fitting these sensorgram data to a mathematical model allows scientists to calculate the association (ka) and dissociation (kd) rate constants and ultimately determine the binding affinity.
In antibody affinity measurement, the antibody to be analyzed is first captured by a high-affinity anti-IgG antibody immobilized on the surface of the sensor chip. A series of concentrations of antigen are then sequentially injected onto the surface of the chip. Based on the sensorgram, mAbs with superior properties can be selected according to kinetic parameters. The identification of specific regions (epitopes) is more promising in drug discovery than the discovery of tight-binding antibodies because binding affinity maturation can be achieved by standard protein engineering. Therefore, based on a bivalent analyte model involving two binding reactions, we are able to provide assays to identify antibodies that bind to two different epitopes. First, a universal antibody (such as rabbit anti-mouse immunoglobulin) is covalently attached to the surface of the chip, and then the primary antibody is captured by the previously attached universal antibody. After the unsaturated sites are blocked, the injected antigen will bind to the primary antibody to form an Ab-Ag complex. Subsequent administration of a second monoclonal antibody for assessment of binding affinity, if the second antibody binds a different individual epitope, a characteristic sensorgram will be observed.
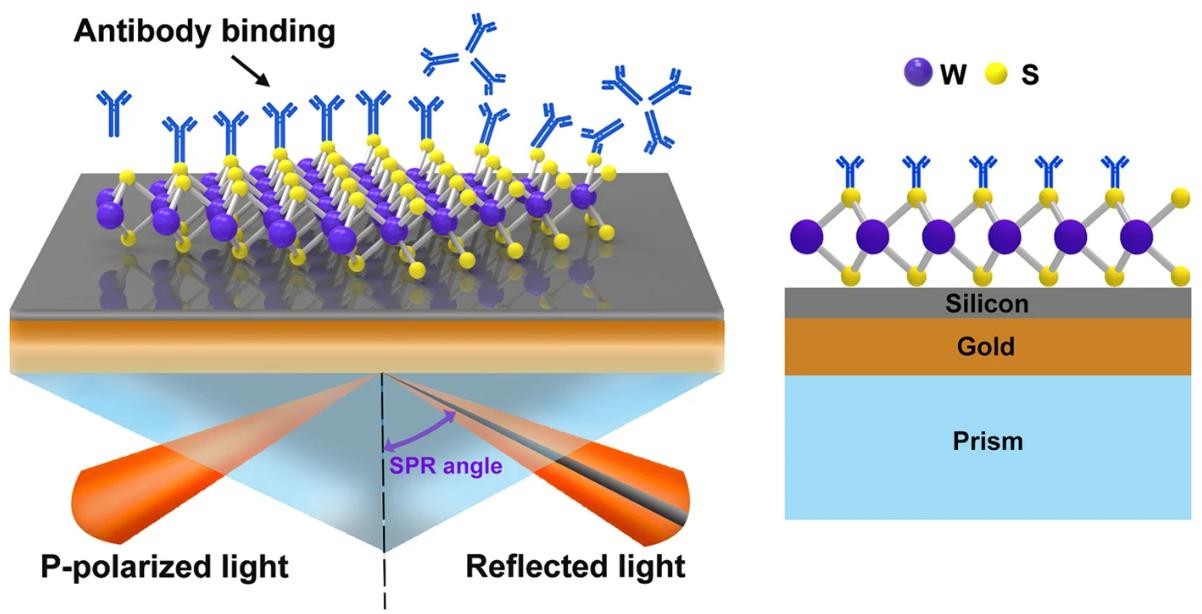 Figure 1. The Principle of Surface Plasmon Resonance. (Ouyang Q, et al. 2016)
Figure 1. The Principle of Surface Plasmon Resonance. (Ouyang Q, et al. 2016)Advantages of an SPR Assay
Label-Free Analysis: SPR eliminates the need for fluorescent or radioactive labeling, preserving the native structure and activity of biomolecules.
Real-Time Kinetics: Observe binding events in real-time to determine association (ka) and dissociation (kd) rates, providing a complete kinetic profile.
Sensitivity: SPR can detect interactions with picomolar affinity, making it ideal for weak and strong binding studies.
Low Sample Volume: Only small quantities of ligand and analyte are required, enabling cost-effective assays even with limited materials.
Flexibility: SPR accommodates a broad range of molecules, including proteins, peptides, nucleic acids, carbohydrates, lipids, and small molecules.
High Throughput: Modern SPR systems support multiplexed assays, accelerating data acquisition for complex interaction networks.
What does SPR Tell Us?
- Interaction specificity
- Interaction affinity
- Kinetic binding parameters
- Hermodynamic parameters
- Biologically active concentration of an analyte
We provide SPR analysis services to study biomolecular interactions. Study a wide variety of biological molecules by SPR, including small molecules (<100 Da), proteins, nucleic acids, lipids, bacteria, viruses, and whole cells. Most published SPR studies involve protein-protein interactions, with antibody-antigen interactions representing a major direction. We provide high-sensitivity instruments and accurate experimental protocols to help customers conduct research on small molecules, lipids and nucleic acids.
SPR in Drug Development
Hit Identification and Affinity Screening
SPR facilitates rapid screening of molecular libraries to identify high-affinity binders. By determining KD values, researchers can prioritize leads for further development.
Kinetic Characterization
Understanding the kinetics of drug-target interactions is critical in drug discovery. SPR provides detailed measurements of ka and kd, enabling the calculation of the residence time—a parameter often linked to drug efficacy and duration of action.
Specificity and Selectivity Studies
SPR can assess cross-reactivity by testing compounds against multiple targets, ensuring the specificity of drug candidates.
Mechanism of Action Studies
SPR elucidates the mode of binding—whether competitive, allosteric, or cooperative—shedding light on a molecule's mechanism of action.
For antibody development, SPR enables detailed epitope mapping, identifying the specific regions of an antigen that bind to the antibody.
Fragment-Based Drug Design
SPR plays a key role in fragment-based approaches, detecting weak interactions that are difficult to measure with other methods.
Sample Requirement for SPR Analysis
| Customer Provided | Requirement |
|---|---|
| Antibodies, proteins, peptides, compounds, small molecules, etc. | -Buffer: PBS, HEPPS, etc. without organic reagents, without Tris -Antigen sample: Protein (peptide) >2mg, concentration >0.5mg/ml, purity >90% -Protein sample (with antibody): >200ug, concentration >0.5mg/ml, purity >90% -Peptide samples: >200ug, concentration >1mg/ml, purity >90% -Compound small molecule: >1mg, concentration >1mg/ml (dry powder is best) |
Demo
Demo 1: Pathogens detection by immunosensing
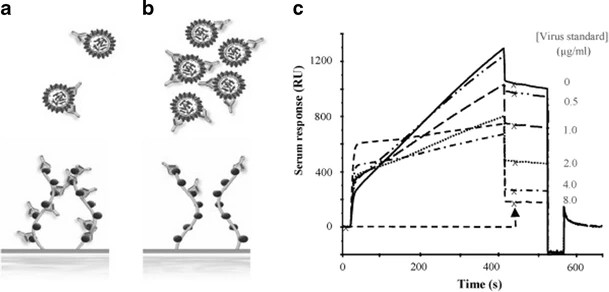 Figure 2. Overlay plot showing sensorgrams of injected serum mixed with a concentration series of virus standard (Nilsson C E, et al., 2010).
Figure 2. Overlay plot showing sensorgrams of injected serum mixed with a concentration series of virus standard (Nilsson C E, et al., 2010).Demo 2: Detecting small-molecule binding
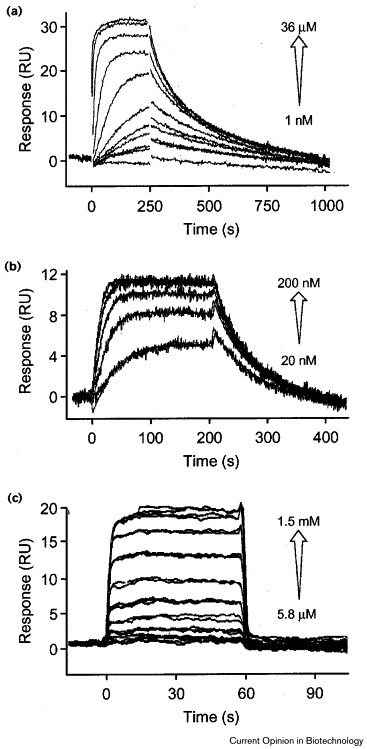 Figure 3. Concentration-dependent analysis of small-molecule–protein interactions. (a) A dihydroxyhexanediamine analog (1 nM–36 μM) binding to immobilized HIV-1 proteinase (Markgren P O, et al., 1998). (b) Novobiocin (molecular mass = 612.7 g/mole) (20–100 nM) binding to an immobilized DNA gyrase truncate (Kampranis S C, et al., 1999). (c) Replicate injections of lactose (molecular mass = 342.3 g/mole) (5.8 μM–1.5 mM) binding to an immobilized antibody (Malmqvist M. 1999).
Figure 3. Concentration-dependent analysis of small-molecule–protein interactions. (a) A dihydroxyhexanediamine analog (1 nM–36 μM) binding to immobilized HIV-1 proteinase (Markgren P O, et al., 1998). (b) Novobiocin (molecular mass = 612.7 g/mole) (20–100 nM) binding to an immobilized DNA gyrase truncate (Kampranis S C, et al., 1999). (c) Replicate injections of lactose (molecular mass = 342.3 g/mole) (5.8 μM–1.5 mM) binding to an immobilized antibody (Malmqvist M. 1999).FAQ
Q: How is immobilization of the ligand on the SPR chip performed?
A: The ligand is immobilized on the sensor chip surface using chemical or physical methods. Common approaches include:
Amine coupling: Covalent binding via primary amine groups.
Biotin-streptavidin binding: Highly specific and strong non-covalent attachment.
Hydrophobic or His-tag binding: Useful for certain proteins or peptides.
Q: Can Creative Proteomics analyze low-affinity interactions?
A: Yes, but within the limitations of the SPR technology. Interactions with a KD in the micromolar to nanomolar range are ideal for SPR. For weaker interactions (e.g., KD in the millimolar range), SPR may require specialized conditions or alternative techniques like isothermal titration calorimetry (ITC). Creative Proteomics provides expert advice on the feasibility of measuring low-affinity interactions.
Q: What types of interactions can SPR measure?
A: SPR is versatile and can analyze a wide range of interactions, including protein-protein, protein-DNA/RNA, small molecule-protein, and even cell-surface interactions.
Case Study
Case: Casein interactions studied by the surface plasmon resonance technique
Background
The study examines the molecular interactions between casein proteins, focusing on how factors like pH, ionic strength, phosphorylation, and calcium affect their binding. Caseins, essential for milk micelle stability and enzymatic coagulation, interact through hydrophobic, electrostatic, and calcium-mediated forces. While previous studies have explored these interactions indirectly, this research utilizes surface plasmon resonance (SPR) technology to offer quantitative insights into casein binding dynamics under controlled conditions. The goal is to deepen the understanding of casein micelle assembly and functionality in biological and industrial contexts.
Methods
Protein Binding Assays Using SPR:
Casein interactions (αs-, β-, and κ-caseins) were studied using immobilized casein molecules on SPR sensor chips.
Binding experiments were conducted at varying pH levels (7.4, 6.4, and 5.4) and ionic conditions to evaluate electrostatic and hydrophobic contributions to casein association.
Effect of Calcium and Ionic Strength:
Binding assays were performed in the presence and absence of 2 mM calcium and 150 mM NaCl to examine their roles in casein interactions.
Role of Phosphorylation:
Dephosphorylated αs-caseins were compared to native αs-caseins to investigate the impact of phosphorylation on calcium-dependent interactions.
Quantifying Hydrophobicity:
SPR technology was applied to measure interactions of caseins on hydrophobic surfaces, and results were correlated with literature values for casein hydrophobicity.
Results
Lowering pH from 7.4 to 5.4 increased casein binding by reducing negative charge and enhancing non-ionic interactions, especially among phosphorylated caseins. At neutral pH, κ-casein showed the highest binding, aligning with its lower electronegativity.
Calcium (2 mM) enhanced casein binding in the absence of NaCl by neutralizing negative phosphate charges and acting as a cross-linker. Binding was strongest for αs- and β-caseins, which have high-affinity calcium-binding sites.
Dephosphorylated αs-caseins bound more strongly to immobilized caseins due to reduced electrostatic repulsion, with calcium effects noticeable only with κ-casein.
Hydrophobicity measurements showed β-casein > κ-casein > αs-caseins, with higher temperature enhancing hydrophobic interactions, crucial for casein aggregation.
Electrostatic repulsions at neutral pH reduced binding, while NaCl reduced repulsion and slightly boosted interactions.
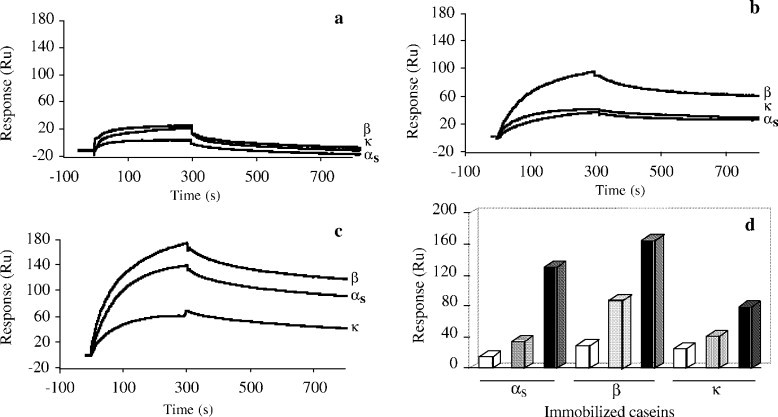 Figure 4. SPR analysis of the interaction between soluble caseins and caseins immobilized on a CM5 sensor chip at 25°C.
Figure 4. SPR analysis of the interaction between soluble caseins and caseins immobilized on a CM5 sensor chip at 25°C.References
- Ouyang Q, et al. Sensitivity enhancement of transition metal dichalcogenides/silicon nanostructure-based surface plasmon resonance biosensor. Scientific reports, 2016, 6(1): 28190. DOI: 10.1038/srep28190
- Nilsson C E, et al. A novel assay for influenza virus quantification using surface plasmon resonance. Vaccine, 2010, 28(3): 759-766. DOI: 10.1016/j.vaccine.2009.10.070
- Markgren P O, Hämäläinen M, Danielson U H. Screening of compounds interacting with HIV-1 proteinase using optical biosensor technology. Analytical biochemistry, 1998, 265(2): 340-350. DOI: 10.1006/abio.1998.2927
- Kampranis S C, et al. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry, 1999, 38(7): 1967-1976. DOI: 10.1021/bi982320p
- Malmqvist M. BIACORE: an affinity biosensor system for characterization of biomolecular interactions. Biochemical society transactions, 1999, 27(2): 335-340. DOI: 10.1042/bst0270335
- Marchesseau S, Mani J C, Martineau P, et al. Casein interactions studied by the surface plasmon resonance technique. Journal of dairy science, 2002, 85(11): 2711-2721. DOI: 10.3168/jds.S0022-0302(02)74358-0
Related Services
Support Documents
KNOWLEDGE CENTER