- Service
- Demo
- Case Study
- FAQ
- Related Services
- Inquiry
Methylation Analysis of proteins is a critical part of biologics development. Our CRO offers comprehensive protein Methylation Analysis to study methylation variants in antibodies and other protein drugs. We use advanced Methylation mass spec methods and workflows to meet regulatory standards. Our workflows comply with ICH Q6B and related guidelines.
What Is Protein Methylation
Protein methylation is a modification where methyl (–CH₃) groups are added to certain amino acids. Methylation can have strong effects on a molecule's properties. For example, adding a methyl group can make a drug stronger and protect it from enzyme breakdown. It can also change how a drug dissolves and how it is taken up by cells. It can change how the body processes the drug. Methylation often occurs on arginine or lysine residues in proteins.
Impacts of Methylation: It can make a drug work better and last longer. Methylation may change drug solubility and how the drug is cleared from the body. These changes can affect the drug's safety or potency. Unexpected modifications can trigger immune responses in patients.
Importance in Drug Development
Protein products often need complete analysis of their structure. Modifications like methylation can influence a protein drug's potency, stability, and safety. Incorrect or variable changes may cause misfolding or immune reactions. Guidelines (ICH Q6B, USP <1047>) require careful analysis of these changes for product quality.
Why Profile Methylation: Confirm the identity and purity of the drug. Ensure consistency and stability across batches. Support regulatory submissions with detailed data. Help guide formulation and comparability studies.
Methylation Analysis Services: Enhancing Precision and Efficiency
At Creative Proteomics, we specialize in delivering top-tier methylation analysis services. Our methods marry expertise with technology, utilizing cutting-edge approaches like high-performance liquid chromatography (HPLC) paired with mass spectrometry (MS) to ensure precision.
Innovative Peptide Enrichment Techniques
Our protein analysis begins by digesting proteins either in gel or solution. Subsequently, we enrich methylpeptides using antibodies finely tuned to identify and target specific methylation patterns. Despite the challenges presented by the minimal alteration in hydrophobicity due to methylation, our blend of chromatography-based techniques, such as strong cation exchange (SCX), isoelectric focusing (IEF), and hydrophilic interaction liquid chromatography (HILIC), maximizes peptide enrichment. The choice of methodology is tailored to align with your project's unique demands, ensuring optimal yield. Post enrichment, our team employs the Orbitrap Fusion Lumos Tribrid Mass Spectrometer for comprehensive MS/MS analysis.
Tailored Metabolic Labeling Strategies
Understanding the significance of protein methylation dynamics, we provide measurement services through advanced metabolic labeling strategies and MS analysis. Our protocols are not one-size-fits-all; they are carefully refined to suit your specific project requirements and goals. This personalized approach enhances the accuracy and reliability of methylation rate assessments.
Our Protein Methylation Analysis Suite
| Service | Description |
|---|---|
| Site Discovery & Mapping | Identify mono-, di-, and tri-methylated lysine/arginine via LC MS/MS |
| Quantitative Profiling | Relative abundance across samples using SILAC, TMT/iTRAQ, or label-free |
| Enzyme Specificity Analysis | Track methyltransferase activity (e.g., PRMTs, KMTs) |
| Stability & Stress Testing | Monitor methylation changes under stress (e.g., temperature) |
| Regulatory Reporting | Full compliance with ICH Q6B, PharmEu, USP <1047> |
Substance Types
We support:
- Purified proteins (≥50 µg)
- Monoclonal antibodies
- Cell lysates, tissues, serum, CSF, PBMCs
- Microbial proteins
Sample Requirements
- Purified protein: ≥50–100 µg
- Cell pellets: 10⁷–10⁸ cells
- Tissue: 100–200 mg fresh/frozen
- Serum/Plasma/CSF: ≥0.5–1 mL
- PBMCs: 5–10 million cells
Methylation Analysis Workflow
Our complete workflow is optimized for protein drugs. We digest the sample into peptides and capture methylated peptides using antibody-based methods. The enriched sample is analyzed by high-resolution LC-MS/MS. We use this to find and measure methylation sites.
Techniques Used:
- Protease digestion of antibodies or proteins.
- Capturing methylated peptides using antibodies or chromatography.
- LC-MS/MS mapping on high-resolution instruments.
- Data analysis with software to assign methylation sites.

Our method can detect both single and double methylation on the same site. We use modern instruments. This gives the best sensitivity and speed.
Advantages of Our Analysis Services
- High Sensitivity and Precision: Capable of detecting even subtle methylation patterns.
- State-of-the-Art Equipment: High-resolution tools provide unmatched sensitivity.
- Optimized Analysis Platform: Our automated chromatographic systems enhance flexibility and reduce analysis time.
- Custom Solutions: Analysis protocols tailored to fit project-specific requirements.
- Experienced Team: Specialists provide expert support from start to finish.
- Efficient Reporting: Clear, customized reports include all relevant details and raw data.
- Quality Assurance: Every result undergoes rigorous quality checks and peer reviews.
At Creative Proteomics, we provide a comprehensive methylation profile of your proteins and antibodies. Our services can be integrated with stability and potency assessments to provide a holistic view of your project's progress.
Demo Results Overview

Case Study
Decoding protein methylation function with thermal stability analysis
Nature Communications, 2023. https://doi.org/10.1038/s41467-023-38863-1
Background
Although methylation is well-studied in epigenetics, its broader functional roles in non-histone proteins remain under-explored . The Sayago study hypothesized that methylation-linked proteins would show detectable shifts in thermal stability—serving as a proxy for functional modulation
Methods
Cell Treatment:
mESCs were treated with PRMT5 inhibitors (GSK591, GSK595) or DMSO control.
Thermal Profiling (PISA):
Heated samples across temperature gradients.
Collected soluble protein fractions at each temperature.
TMT Labeling & LC MS/MS:
Samples labeled using TMTpro reagents.
Peptides analyzed via Orbitrap instruments.
Thermal Stability & Abundance Analysis:
Quantified stability shifts and protein abundance via TMT-based MS.
Multiple biological replicates ensured statistical robustness.
Data Analysis:
Identified proteins with altered melting curves indicating methylation-linked changes, independent of abundance.
Key Findings
PRMT5 Regulates mRNA Binding Proteins:
Thermal profiles revealed destabilization of proteins rich in intrinsically disordered regions.
These proteins are implicated in liquid-liquid phase separation and stress granule formation.
Chaperonin (TRiC/CCT) Subunits:
Eight subunits exhibited stability shifts upon PRMT5 inhibition—confirming known Prmt5 substrates.
Ezh2 Non-Canonical Function:
Ezh2, conventionally known for histone methylation, showed altered thermal stability within mitotic chromosomes and perichromosomal regions.
Mki67 was identified as a novel Ezh2 substrate.
Disconnection Between Abundance and Stability:
Several proteins showed stability shifts without abundance changes, highlighting the assay's sensitivity to post-translational modulation .
 Thermal proteome analysis of mESCs upon inhibition of Prmt5.
Thermal proteome analysis of mESCs upon inhibition of Prmt5.Conclusions
Functional Readout of Methylation: Thermal stability changes reveal active methylation-driven functional shifts beyond what abundance alone shows.
Discovery Potential: Highlights previously unknown methylation roles in chromatin and mitotic regulation (e.g., Ezh2, Mki67) .
Screening Utility: Validates PISA as a scalable, unbiased tool for functional methylomics profiling.
FAQ
Q: How much sample is required?
- Purified proteins: ≥50 µg
- Cell lysates: ≥10⁷ cells
- Tissue: ≥100–200 mg
- Biofluids: ≥0.5–1 mL
Q: Can we detect histidine methylation?
A: Yes. With methyl-specific labeling and enzymes like CARNMT1 we have validated histidine methylation detection.
Q: How do you avoid false positives?
- Enrichment via antibody panels plus HILIC/SCX
- Orthogonal heavy SILAC reduces false discovery
Q: Is quantitation possible?
A: Yes:
- SILAC, TMT/iTRAQ, or label-free
- Detect fold-changes across conditions
Q: Will data support regulatory filings?
A: Absolutely. We prepare full compliance documents aligned with ICH Q6B and USP <1047>.
References
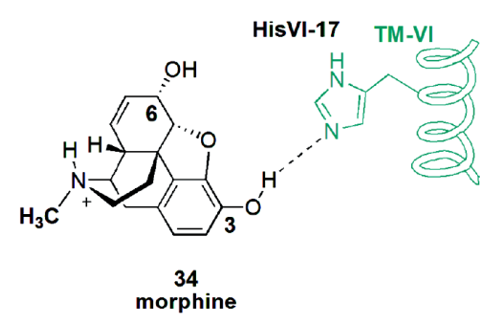
- Barreiro E.J., Kummerle A.E., and Fraga C.A.M., The methylation effect in medicinal chemistry. Chem. Rev. 2011, 111, 5215–5246. DOI: 10.1021/cr200060g
- Sun S. and Fu J., Methyl-containing pharmaceutical: Methylation in drug design. Bioorganic & Medicinal Chemistry Letters 28 (2018) 3283–3289. DOI: 10.1016/j.bmcl.2018.09.016
- Wang Q., Wang K., Ye M. Strategies for large-scale analysis of non-histone protein methylation by LC-MS/MS. Analyst. 2017 Sep 25;142(19):3536-3548. DOI: 10.1039/c7an00954b
Related Services