- Services
- Demo
- FAQ
- Case Analysis
- Related Services
Detect, Characterize, and Control Protein Aggregates Across Biologic Development Pipelines
Protein aggregation is one of the most critical challenges in biologic drug development. Even low levels of aggregates can compromise therapeutic safety, potency, and regulatory approval timelines. At Creative Proteomics, we provide a comprehensive protein aggregation assay portfolio—designed to pinpoint the root causes of aggregation, quantify monomer loss, and deliver the insights you need to optimise your molecule and process.
Whether you are advancing a monoclonal antibody, an antibody–drug conjugate, or a recombinant protein, our team combines orthogonal testing platforms and regulatory expertise to ensure your product meets global quality standards.
Why Protein Aggregation Analysis Matters
In biologics development, protein aggregation isn't just a lab artefact—it's a regulatory and patient safety concern. Aggregates can form during cell culture, purification, formulation, filling, storage, and transport. Even minor levels of aggregation can alter a drug's behaviour in ways that impact its performance and approval readiness.
- Loss of potency: Aggregates reduce the active monomer fraction, weakening therapeutic efficacy.
- Increased immunogenicity risk: Abnormal protein aggregation can trigger anti-drug antibodies that neutralise treatment—or worse, cross-react with endogenous proteins.
- Regulatory implications: Authorities view aggregate levels as a Critical Quality Attribute (CQA). Uncontrolled aggregation can delay or derail IND or BLA submissions.
Creative Proteomics partners with biopharma teams to detect, quantify, and interpret aggregates before they compromise quality or compliance.
Causes of Protein Aggregation
Protein aggregation can begin the moment a biologic molecule is expressed—and continue through every stage of development. Understanding the root causes is essential for designing effective protein aggregation assays and mitigation strategies.
Structural Drivers
- Hydrophobic patches: Exposed regions on antibody frameworks or recombinant proteins often seed aggregation.
- Glycosylation loss: Changes in sugar moieties, especially in the CH2 domain of antibodies, destabilise the Fc region and accelerate aggregate formation.
- Unpaired thiols: Free sulfhydryl groups can crosslink, forming covalent aggregates.
Process & Manufacturing Stresses
- Upstream events: Aggregates may form during cell culture when proteins misfold or when molecular chaperones fail to stabilise new chains.
- Downstream factors: Harsh purification conditions (e.g. low pH Protein A elution) or repetitive freeze–thaw cycles increase aggregation.
- Mechanical stress: Stirring, pumping, or filtration can expose proteins to shear forces that destabilise structure.
Environmental Triggers
- pH and ionic strength: Proteins are most aggregation-prone near their isoelectric point.
- Temperature: Heat accelerates chemical degradation and exposes hydrophobic cores.
- Trace metals or excipient impurities: Can nucleate aggregation or destabilise formulations.
Our Protein Aggregation Assay Services Portfolio
We use orthogonal testing platforms to deliver a complete, reliable picture of protein aggregation—from nanometre-scale oligomers to visible particles.
SEC-MALS (Size-Exclusion Chromatography with Multi-Angle Light Scattering)
- Separates monomers, dimers, and trimers by size.
- MALS detector provides absolute molar mass, eliminating reliance on calibration standards.
- Ideal for routine monitoring during process development and formulation screening.
SV-AUC (Sedimentation Velocity Analytical Ultracentrifugation)
- A column-free orthogonal method that regulators favour for aggregate characterisation.
- Detects reversible and irreversible aggregates without dilution artefacts.
DLS (Dynamic Light Scattering)
- Rapidly screens for early-stage aggregate formation and size distribution in solution.
- Particularly useful for formulation stress tests and forced degradation studies.
MFI (Micro-Flow Imaging) & Light Obscuration
- Detect and image sub-visible particles in the 2–100 µm range, aligned with USP <787>/<788> requirements.
- Distinguish proteinaceous particles from silicone oil droplets or other contaminants.
Spectroscopy (CD, FTIR, nUV)
- Tracks changes in secondary and tertiary structure that precede aggregation.
- Provides insight into unfolding events or excipient-driven stabilisation.
Optional & Supporting Methods
- Turbidity and visual inspection for quick checks of visible aggregates.
- Calorimetry (DSC) for evaluating thermal stability and unfolding profiles.
What are the methods of protein aggregation?
| Method | What It Detects | Sample Needs |
|---|---|---|
| SEC-MALS | Monomers, dimers, trimers; absolute molar mass | ~50 µL per run; serum, plasma, or formulation buffer |
| SV-AUC | Reversible & irreversible aggregates; sedimentation profile | ~150 µL; minimal buffer constraints |
| DLS | Early-stage aggregates & size distribution (1–1000 nm) | 10–20 µL; suitable for formulation screening |
| MFI / Light Obscuration | Sub-visible particles (2–100 µm); morphology data | 200–500 µL; clear solutions preferred |
| Spectroscopy (CD, FTIR, nUV) | Secondary & tertiary structure shifts | 50–100 µL; requires optically clear samples |
| Calorimetry (DSC) | Thermal stability & unfolding temperature (Tm) | ≥200 µL; protein concentration >0.5 mg/mL |
| Turbidity / Visual Inspection | Visible particulates & opalescence | Minimal; routine QC samples |
How Our Process Works
Our protein aggregation analysis is designed to be clear, efficient, and aligned with your project goals. From first contact to final report, every step is built for reliability and transparency.
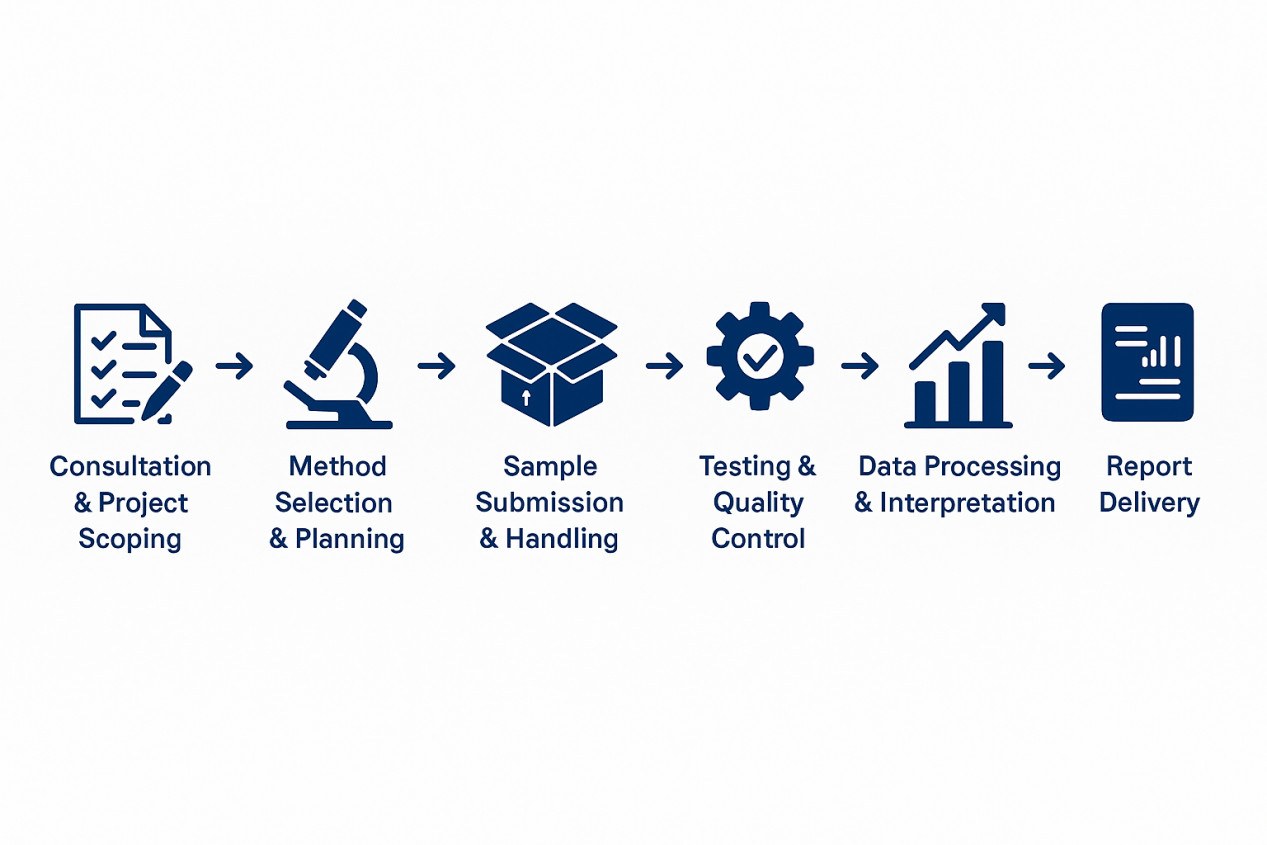
Deliverables & Reporting
- Aggregate profile summary – monomer, oligomer, and particle breakdown
- Raw and processed data files – chromatograms, AUC plots, imaging outputs
- Visual reports – graphs, tables, and particle morphology images
- QC documentation – method performance and control checks
- Regulatory-ready package – ICH Q6B-aligned interpretations for submissions
- Optional consultation – review meeting to discuss findings and next steps
Why Choose Creative Proteomics?
- Orthogonal expertise: Integrated SEC-MALS, SV-AUC, DLS, and MFI workflows for complete coverage.
- Regulatory alignment: Data packages meet ICH Q6B standards and support IND/BLA filings.
Rapid turnaround: Most projects completed in 2–3 weeks with flexible scheduling.
- Experienced scientists: 10+ years in biologics QC and aggregation analysis.
- Global reach: Coordinated labs and project managers across the U.S., EU, and Asia.
- End-to-end support: From early-stage discovery to preclinical comparability and stability studies.
Applications of Protein Aggregation Analysis
Developability assessment – Identify aggregation risks early during candidate screening to guide molecular design decisions.
Comparator studies – Benchmark biosimilars against originator products by comparing aggregate profiles.
Formulation development – Screen buffers, excipients, and pH conditions to minimize aggregation in final drug product.
Stability testing – Track aggregate formation under real-time and accelerated stability conditions.
Demo
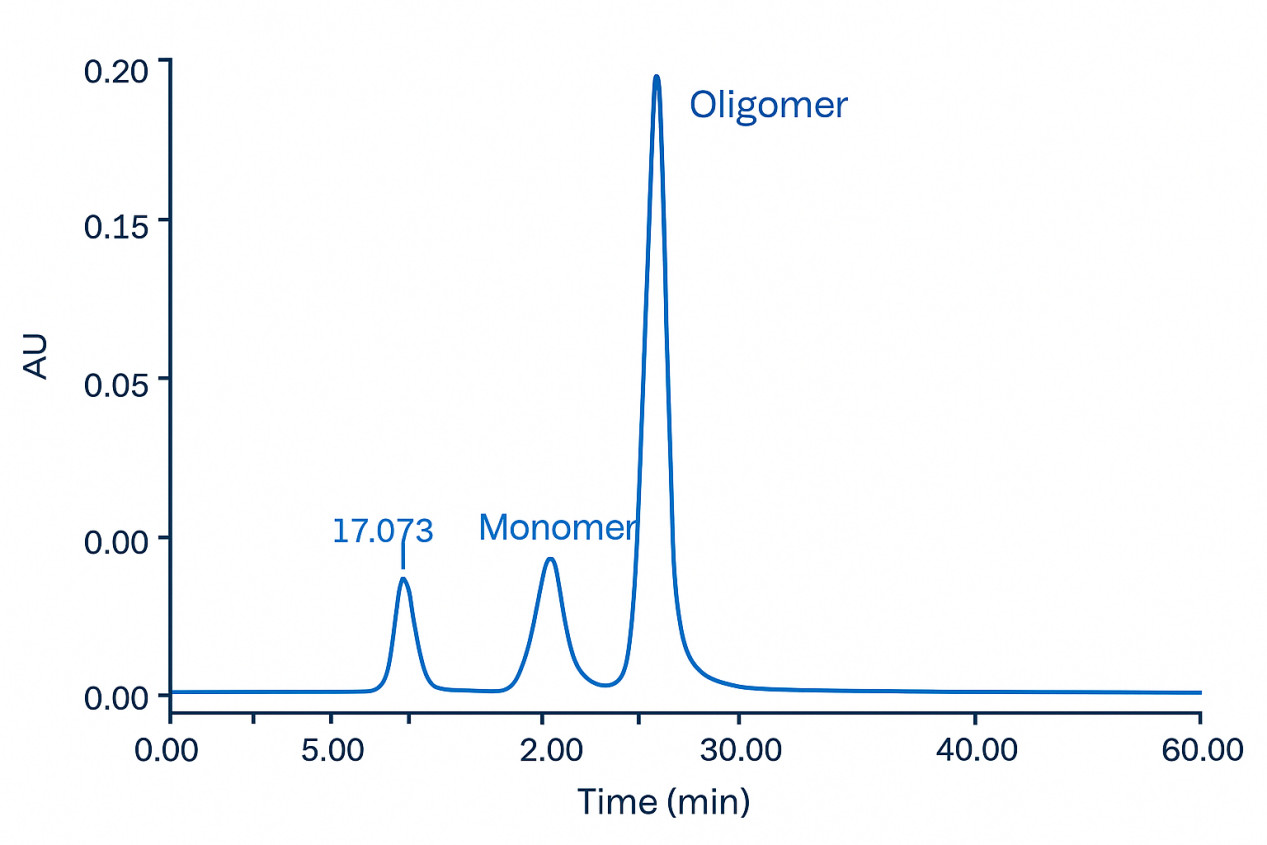 SEC-HPLC chromatogram showing the separation of human serum albumin oligomer, dimer, and monomer.
SEC-HPLC chromatogram showing the separation of human serum albumin oligomer, dimer, and monomer.FAQs – Protein Aggregation Analysis
What is an aggregation assay?
An aggregation assay is an analytical test designed to detect and quantify protein aggregates in biologic drugs, from small oligomers to visible particles.
How does a protein assay work?
Protein assays detect physical or chemical changes in a protein—such as size, shape, or structure—using techniques like chromatography, spectroscopy, and light scattering.
What is the process of protein aggregation?
Protein aggregation begins when proteins misfold or unfold, exposing hydrophobic patches that stick together, eventually forming dimers, oligomers, or insoluble particles.
What is abnormal protein aggregation?
Aggregation that changes a drug's potency, stability, or safety profile—especially when unexpected or excessive—is considered abnormal and requires investigation.
What is protein aggregation?
Protein aggregation is the clustering of protein molecules into larger complexes—covalent or non-covalent—that can compromise product quality or trigger unwanted immune responses.
How do you determine the aggregation of a protein?
Our assays combine orthogonal methods—such as SEC-MALS, SV-AUC, and micro-flow imaging—to provide a complete picture of aggregation at every stage of your process.
Why do I need more than one aggregation method?
Regulators expect orthogonal testing. A single method can miss certain aggregates; combining techniques ensures comprehensive detection (e.g. non-covalent aggregates invisible to one platform).
Case Analysis
Rational Engineering to Reduce mAb Aggregation
Background
Protein aggregation remains a critical hurdle in monoclonal antibody (mAb) development—impacting manufacturability, stability, safety, and immunogenicity. van der Kant et al. (2017, Journal of Molecular Biology) explored aggregation-prone regions (APRs) and engineered "aggregation gatekeeper" mutations to mitigate these risks while preserving antigen binding.
Methods
- In silico APR identification: Using TANGO for intrinsic aggregation propensity and FoldX for local thermodynamic stability (ΔGcontrib), researchers mapped exposed critical APRs in mAb CDRs and framework regions.
- Rational mutagenesis: Introduced charged or proline residues ("gatekeepers") to reduce aggregation propensity without compromising structural stability.
- Orthogonal experimental validation: Engineered variants and wild-type controls were tested via SEC, DLS, and DSC/DSC-Tagg measurements to confirm aggregation reduction.
Results
APR engineering resulted in variants with significantly reduced aggregation propensity. One variant (SL50K + FH101P) increased Tagg by ~15 °C, effectively eliminating aggregation under native conditions.
Improved expression titer: Up to 4.5-fold higher yield compared to wild type, offering direct manufacturability benefits.
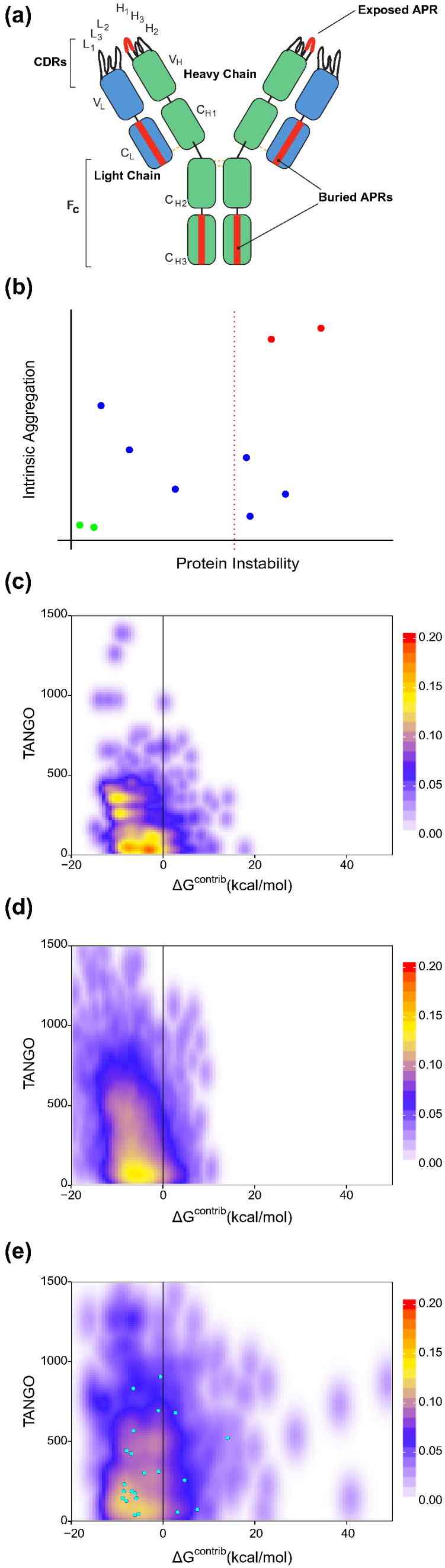 Fig 1. Stretch-plot showing aggregation propensity (TANGO score) versus local stability (ΔGcontrib) of antibody APRs; critical APRs clustered in top-right indicate aggregation risk under native conditions.
Fig 1. Stretch-plot showing aggregation propensity (TANGO score) versus local stability (ΔGcontrib) of antibody APRs; critical APRs clustered in top-right indicate aggregation risk under native conditions.Conclusion
- Demonstrates how early structural prediction combined with orthogonal aggregation assays (SEC-MALS, SV-AUC, DLS, MFI) can guide actionable molecule design.
- Highlights the importance of targeting **critical APRs—especially within CDRs—to improve stability, yield, and reduce immunogenic risk.
- Validates our integrated workflow: pairing in silico APR prediction with multi-method aggregation profiling, to support candidate selection, process development, and formulation optimization.
Related Services