- Backgrourd
- Service
- FAQ
- Demo
- Case Study
- Related Services
- Support Documents
- Inquiry
What Is Amino Acid Composition Analysis?
Amino acid composition analysis is a core technique in protein science that determines the relative or absolute abundance of amino acids within a protein or peptide sample. The method typically involves complete acid hydrolysis of the protein into its constituent amino acids, followed by quantitative separation and detection using chromatographic systems such as HPLC or UPLC. Each amino acid is measured independently, and the resulting data provides a precise molecular "fingerprint" of the protein's composition.
This analytical approach is indispensable for protein characterization, as it supports validation of primary structure, confirms theoretical sequences, and helps detect anomalies such as truncations or modifications. Importantly, because it does not require prior sequence knowledge, amino acid composition analysis is especially valuable in early-stage protein evaluation, quality control, and formulation development workflows.
The information obtained from this analysis also serves as a foundation for regulatory documentation, especially when characterizing biologics, biosimilars, or recombinant proteins. For research teams in biopharma or academic settings, it provides a reliable method to verify protein integrity, purity, and consistency across development pipelines.
Why Amino Acid Composition Analysis Matters in Biologics
In the development and manufacture of biotherapeutics—such as monoclonal antibodies, fusion proteins, and recombinant enzymes—amino acid composition analysis plays a vital role in ensuring product consistency, identity, and structural integrity. For both pharmaceutical R&D and contract research organizations (CROs), this technique offers a powerful tool to validate protein-based products across research, preclinical, and process development stages.
One of the primary applications is confirming protein identity. By comparing the experimentally determined amino acid profile against theoretical values derived from known sequences, researchers can verify the correctness of protein constructs and detect unexpected deviations, such as truncations, substitutions, or impurities. This is particularly important when working with complex expression systems or engineered proteins, where transcriptional or translational errors can alter final composition.
Equally important is the role of amino acid analysis in batch-to-batch consistency assessments. In biomanufacturing, even minor variations in upstream or downstream processes can affect protein expression or degradation profiles. Regular amino acid composition checks help CROs and quality control teams monitor manufacturing consistency and ensure alignment with critical quality attributes (CQAs) defined during development.
Additionally, this method is often used to detect post-translational modifications (PTMs) or degradation events. For instance, partial oxidation, deamidation, or fragmentation may alter amino acid abundance, providing early warning signs of instability. When coupled with orthogonal techniques, amino acid profiling supports comprehensive protein structure verification, helping researchers troubleshoot and optimize expression constructs or storage conditions.
Service Content: What's Included in Our Amino Acid Composition Analysis
Creative Proteomics provides an efficient, rapid, and accurate amino acid composition analysis service tailored to meet the rigorous demands of protein-based drug development and biopharmaceutical quality control. Our workflow aligns with international guidelines, including ICH Q6B for the characterization of biotechnological products, ensuring consistency, reliability, and data integrity across all projects.
We Can Provide—But Not Limited To:
Amino Acid Identification: Qualitative profiling of standard and select non-standard amino acids within protein and peptide samples.
Amino Acid Quantitation: Precise determination of absolute concentrations and molar ratios using validated calibration standards and internal references.
Workflow
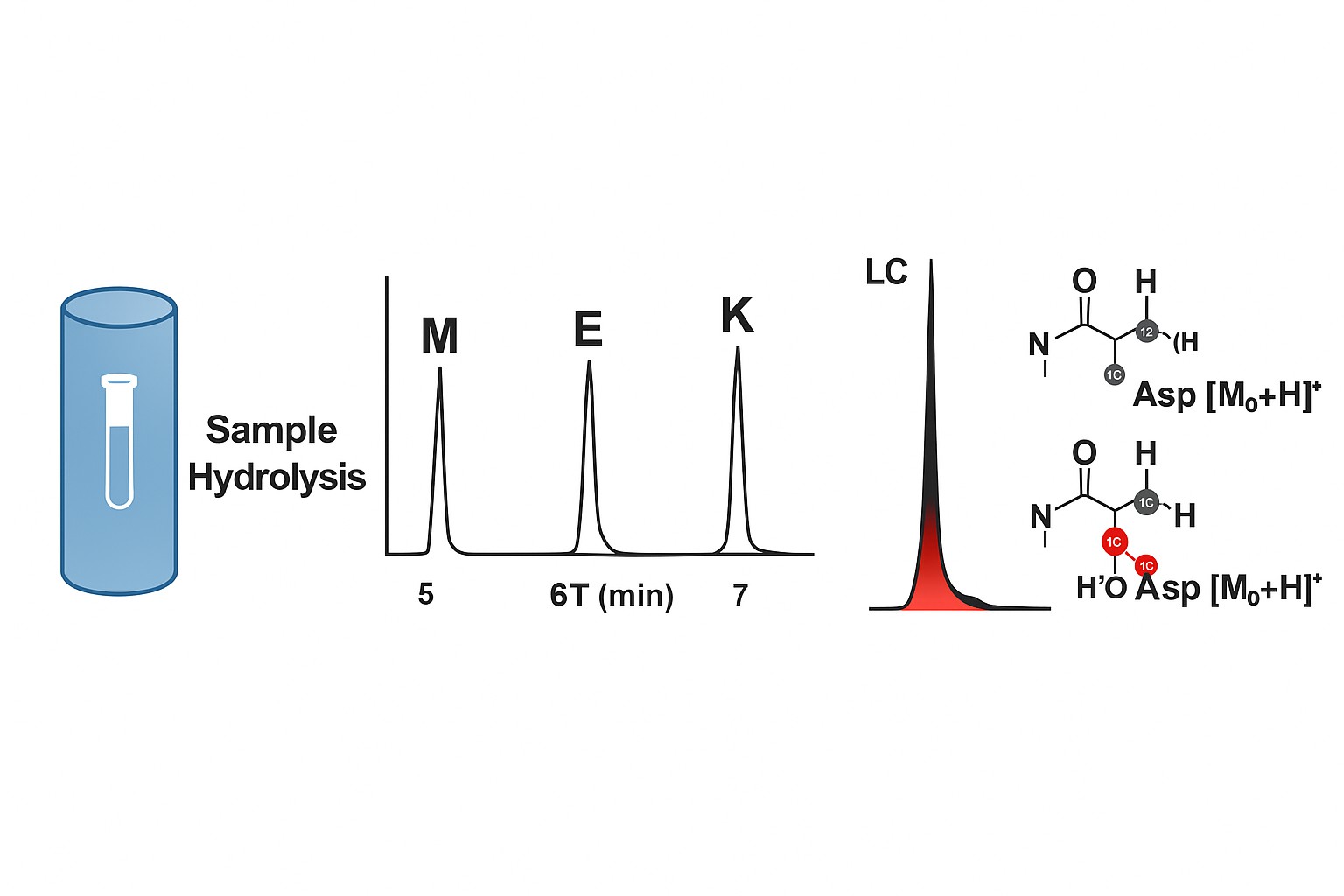
1. Protein Hydrolysis
We utilize 6 M hydrochloric acid (HCl) for complete acid hydrolysis of protein samples under vacuum or nitrogen to minimize oxidation. Additional options include:
- Performic acid oxidation for cysteine/cystine determination
- Alkali hydrolysis for preserving tryptophan
- Enzymatic pre-treatment, if applicable
2. Derivatization Techniques
To enable sensitive detection and high-resolution separation, we offer:
- PITC derivatization (Edman-type) for classical PTC amino acid analysis
- OPA or FMOC derivatization for fluorescence detection of primary and secondary amines
3. Chromatographic Quantification via HPLC or UPLC
Samples are processed on advanced HPLC or UPLC platforms, allowing robust separation and reproducible quantification of amino acids with detection limits down to 1–10 pmol. All measurements are benchmarked against certified amino acid standards.
4. Optional Detection of Modified or Rare Amino Acids
Upon request, we offer specialized detection for:
Hydroxyproline (collagen analysis)
Cysteic acid, methionine sulfone (oxidative stability testing)
Other non-proteinogenic amino acids, depending on sample type
Comprehensive Analytical Report
Each project is delivered with a customized, publication-ready report containing:
Identification and quantification results
Molar ratio profiles and total amino acid content
Method summary, chromatograms, and system suitability data
Notes on rare amino acid detection or sample-specific deviations
This end-to-end service supports both early-stage discovery and late-stage product characterization in accordance with global regulatory expectations.
Our Analytical Platform and Methodologies
Separation Technologies
- Hydrophilic Interaction Liquid Chromatography (HILIC) for efficient separation of underivatized or polar amino acids
- (Ultra) High-Performance Liquid Chromatography (HPLC/UPLC) for PITC- or OPA-labeled amino acids
- Gas Chromatography (GC) and Capillary Electrophoresis (CE) in specialized applications
To mitigate limitations associated with classical derivatization (e.g., instability of fluorescent tags or degradation during GC-MS), we rely on HILIC–tandem mass spectrometry (MS/MS) as our primary detection method. This MS-based platform offers superior sensitivity, eliminates fluorescence interference from endogenous compounds, and avoids the degradation artifacts often seen in GC-based workflows.
Tandem MS Detection and Platform
Our core analytical systems include:
- Prominence-xR UFLC (Shimadzu) for rapid, high-resolution liquid chromatography
- 5500 QTRAP LC-MS/MS (SCIEX) for sensitive, quantitative detection of amino acids via multiple reaction monitoring (MRM)
This HILIC–MS/MS workflow is particularly well-suited for the compositional analysis of biopharmaceutical samples, enabling the accurate detection and quantification of all standard amino acids, as well as oxidized and modified variants, with minimal interference and high recovery rates.
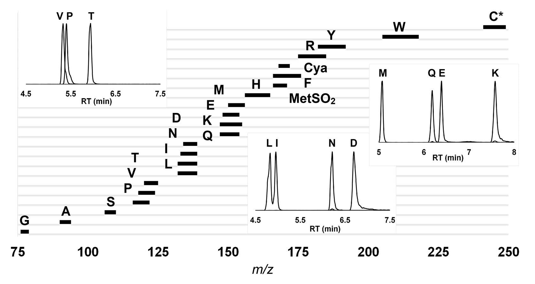 Distribution of amino acids based on their m/z values and retention time.
Distribution of amino acids based on their m/z values and retention time.This figure shows the unique mass-to-charge (m/z) values of amino acids analyzed by tandem mass spectrometry. Each black bar represents the detection window of a specific amino acid derivative. Insets show representative LC chromatograms of selected amino acids (e.g., Val, Glu, Lys) with corresponding retention times under HILIC-MS conditions.
Automated Workflow and Data Integrity
All workflows are automated where possible—from derivatization to injection—to reduce analyst-induced variability and ensure batch-to-batch reproducibility. Advanced software tools are used for peak identification, internal standard calibration, and molar ratio calculation, and every dataset is reviewed by experienced analysts.
Compatible Sample Types and Requirements
| Category | Details |
|---|---|
| Accepted Sample Types | - Purified proteins (e.g., monoclonal antibodies, enzymes, recombinant proteins) - Peptides (synthetic or biologically derived) - Protein formulations (with or without excipients) - Cell lysates or fermentation supernatants (after cleanup) |
| Minimum Sample Amount | ≥ 50 µg of total protein per analysis |
| Recommended Concentration | Ideally ≥ 0.5 mg/mL |
| Buffer Compatibility | Preferably volatile/simple buffers (e.g., ammonium bicarbonate, phosphate) |
| Avoid These Components | Tris, SDS, high glycerol, EDTA, or strong detergents/reducing agents |
| Submission Guidelines | - Label each sample clearly with a unique ID - Provide formulation and concentration details - Note if rare or modified amino acids are expected |
Technical Advantages of Our Service
High Sensitivity and Low Detection Limits
Our amino acid quantification platform can reliably detect amino acids at levels as low as 1–10 picomoles, enabling accurate analysis of scarce or low-abundance samples such as early-stage recombinant proteins, rare peptides, or partially purified formulations. This sensitivity is achieved through tandem mass spectrometry (LC-MS/MS) and refined sample preparation protocols.
Exceptional Reproducibility
We maintain coefficient of variation (CV) values below 5% across technical replicates, ensuring consistency and confidence in batch-to-batch comparisons, biosimilar equivalence studies, and process validation workflows. All datasets are manually reviewed and verified by experienced analysts prior to reporting.
Broad Amino Acid Coverage
Our workflows allow quantitative profiling of 20+ standard and non-standard amino acids.
This broad detection range enhances our capacity to identify PTMs and monitor protein degradation or oxidative stress effects under formulation, storage, or process conditions.
Customized Protocols Based on Sample Type
We tailor hydrolysis, derivatization, and separation strategies based on your specific sample type, target analytes, and downstream applications. Whether working with PEGylated proteins, antibody-drug conjugates, or stress-exposed biotherapeutics, our scientists will recommend optimized protocols to ensure maximum data quality and interpretability.
Rapid Turnaround Time
While project timelines may vary based on sample complexity and batch size, we aim to deliver comprehensive reports within a few working days of receiving qualified samples. Expedited options are available upon request.
Note: All data generated are suitable for internal R&D, CRO-regulated studies, or preclinical documentation, but not for clinical or diagnostic use.
Application Scenarios
1. Therapeutic Protein Lot Release Testing
Amino acid analysis is routinely employed to verify the identity and purity of biopharmaceutical products before release. It provides a quantitative check of the protein backbone against reference values, ensuring that critical quality attributes (CQAs) are maintained throughout production cycles.
2. Biosimilarity Comparability Assessments
For developers of biosimilars, demonstrating equivalence to the reference biologic is a regulatory requirement. Amino acid composition data is a key component of comparability protocols, confirming that the primary structure aligns with the innovator molecule and detecting any sequence discrepancies or formulation-induced degradation.
3. Protein Degradation Profiling
Under stress conditions such as heat, pH shifts, or oxidation, proteins can degrade into fragments or undergo modifications. Amino acid analysis helps characterize these changes by revealing shifts in amino acid ratios, which may signal partial hydrolysis, oxidation (e.g., Met to Met sulfone), or chemical modification.
4. CRO-Based Quality Control of Expression Systems
CROs and core facilities frequently use amino acid composition analysis to validate expression products in early-stage development. Whether expressing recombinant proteins in E. coli, yeast, or mammalian cells, this method ensures the correct product was obtained before investing in downstream purification or scale-up.
5. Validation of Protein Modifications and Conjugates
In studies involving PEGylation, fusion tags, or PTMs, amino acid analysis can confirm successful modification or reveal unintended chemical alterations. It provides orthogonal confirmation to mass spectrometry and SDS-PAGE.
This versatility makes amino acid composition analysis an essential tool across the R&D and preclinical development spectrum.
FAQ – Common Technical and Project Questions
Q: Can you analyze glycoproteins or PEGylated proteins?
A: Yes. While glycosylation or PEGylation can affect protein solubility or hydrolysis efficiency, we routinely analyze glycoproteins and PEG-modified proteins. Please inform us of any conjugations in advance, as additional pre-treatment or cleanup steps may be needed to ensure accurate amino acid recovery.
Q: Do you offer tryptophan-specific analysis?
A: Absolutely. Tryptophan is labile under standard acid hydrolysis conditions and is typically degraded. To address this, we offer alkaline hydrolysis protocols (e.g., NaOH-based), which preserve tryptophan and allow its selective quantification.
Q: Is your method suitable for regulatory documentation?
A: Yes, for research and preclinical purposes. Our workflows follow ICH Q6B-aligned procedures and are suitable for non-GMP, research-use-only documentation. We provide full method details, traceability, and system suitability data with every report.
Q: How much sample do I need to submit?
A: We typically require ≥ 50 µg of total protein at a concentration of ≥ 0.5 mg/mL. For low-abundance samples or complex matrices, we can advise on concentration strategies or perform cleanup steps to meet assay compatibility.
Q: What is the turnaround time?
A: Turnaround time depends on sample type, batch size, and analysis complexity. We aim to deliver results as efficiently as possible without compromising data quality. For time-sensitive projects, expedited options are available. Please contact our team for an estimated timeline based on your specific project requirements.
Q: Can you help interpret the results?
A: Yes. Every project includes a dedicated analyst who can assist with data interpretation, sequence comparison, and integration of amino acid results into broader protein characterization workflows.
If your question isn't listed here, our technical team is always available to provide guidance on custom workflows, special sample types, or regulatory alignment.
Demo
Case Study: Amino Acid Composition Analysis for Monoclonal Antibody Reference Material Characterization
Background
As the development of monoclonal antibody (mAb) therapeutics accelerates, regulatory agencies and pharmaceutical developers require highly reliable reference materials to validate analytical methods and support inter-laboratory comparability. In a study led by the National Metrology Institute of Japan (NMIJ), researchers developed and characterized a monoclonal antibody reference material, AIST-MAB, using amino acid composition analysis as a central method for value assignment.
Objective
The goal was to assign an accurate protein content value to AIST-MAB by quantitatively analyzing its amino acid composition using isotope dilution mass spectrometry (ID-MS), in line with best practices for reference material certification.
Methods and Results
The antibody sample was hydrolyzed using both liquid-phase and gas-phase acid hydrolysis.
Amino acids released from the hydrolysis were quantified using liquid chromatography-tandem mass spectrometry (LC-MS/MS).
The combined results from both hydrolysis techniques yielded a final assigned protein concentration of 5.00 ± 0.19 g/L (expanded uncertainty, k=2; ~95% confidence level).
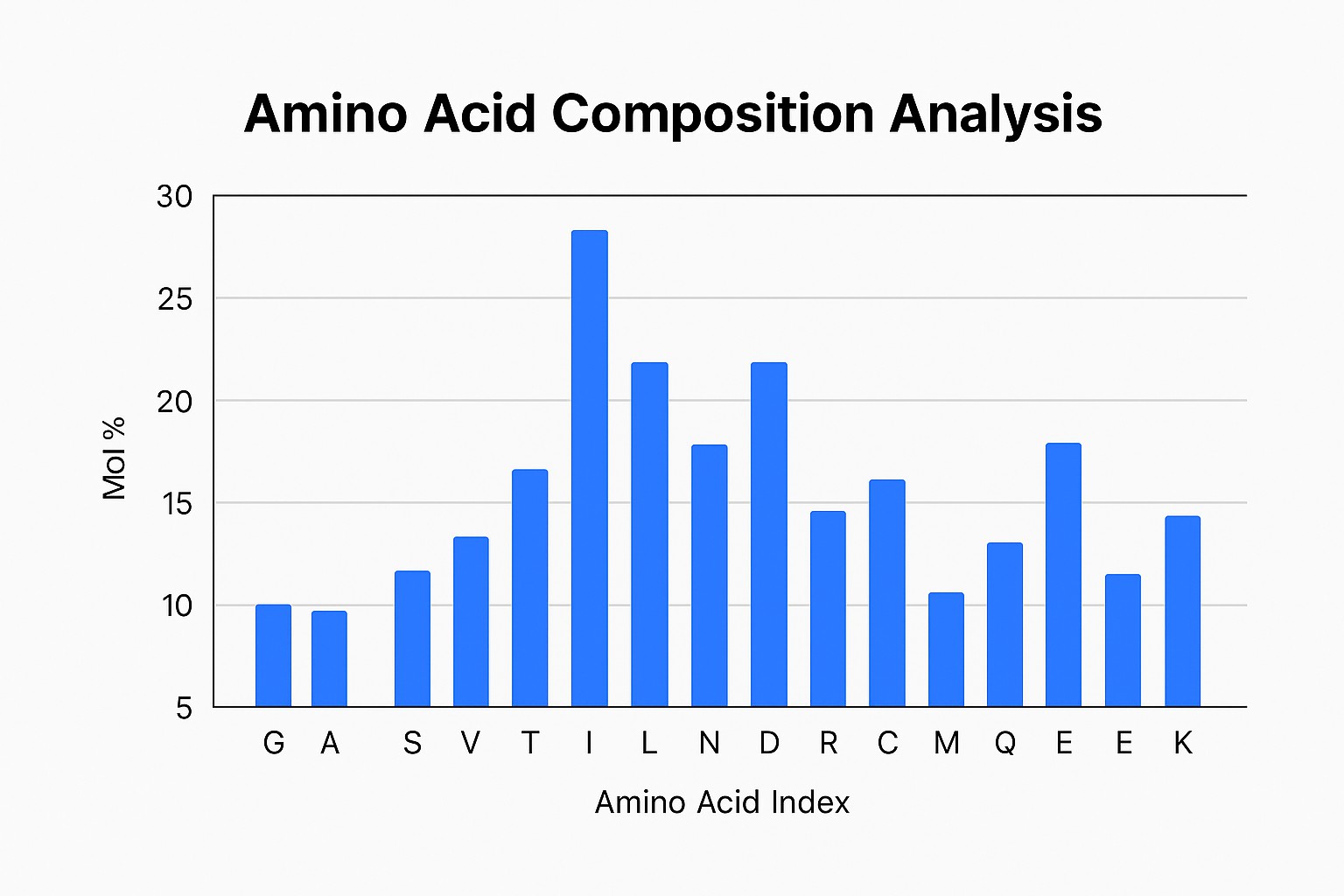
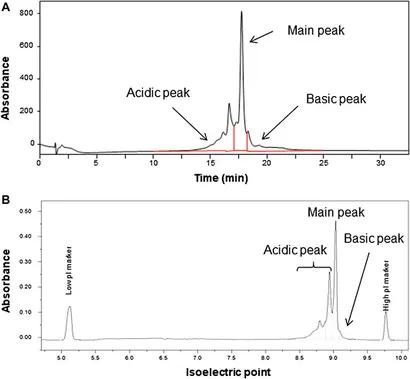 Figure 3 – Quantification results of amino acids used in the ID-MS analysis.
Figure 3 – Quantification results of amino acids used in the ID-MS analysis.Significance
This study demonstrated how amino acid composition analysis—particularly when combined with ID-MS and robust hydrolysis protocols—can be used for the accurate value assignment of complex protein therapeutics. The reference material (AIST-MAB) developed through this process now supports method validation, performance benchmarking, and regulatory compliance in biopharmaceutical analysis.
References
- Kinumi, T., et al. (2022). Characterization and Value Assignment of a Monoclonal Antibody Reference Material. Frontiers in Molecular Biosciences, 9, 842041. DOI: https://doi.org/10.3389/fmolb.2022.842041
Related Services
Support Documents