- Backgrourd
- Services
- FAQ
- Case Study
- Related Services
- Support Documents
- Inquiry
Protein phosphorylation is one of the most common and biologically significant post-translational modifications (PTMs) in eukaryotic cells. As a reversible chemical change catalyzed by protein kinases and removed by phosphatases, phosphorylation plays a crucial regulatory role in cellular processes such as signal transduction, protein stability, enzyme activity, and subcellular localization.
At Creative Proteomics, we offer high-resolution, research-use-only (RUO) Phosphorylated Protein Analysis Services to help biopharmaceutical R&D teams characterize protein phosphorylation patterns across monoclonal antibodies (mAbs), fusion proteins, and recombinant biologics. Our integrated workflow combines high-end mass spectrometry with optimized phosphopeptide enrichment and bioinformatics, supporting both global discovery and targeted quantification.
Why Protein Phosphorylation Matters in Modern Biopharmaceuticals
Protein phosphorylation—the reversible addition of a phosphate group to serine, threonine, or tyrosine residues—is far more than a textbook signaling mechanism. In the biopharmaceutical arena, it is a Critical Quality Attribute (CQA) that can alter a therapeutic protein's conformation, charge state, aggregation propensity, and ultimately its biological function. Even when a drug is intended to bind an extracellular target, subtle lot-to-lot shifts in phosphorylation levels have been shown to modulate potency, stability, and immunogenic risk during pre-clinical evaluation.
Regulatory and Development Implications
Global guidance documents such as ICH Q6B and the FDA's CMC expectations explicitly require characterization and monitoring of PTMs, including phosphorylation, throughout the product lifecycle. Due to the labile and sub-stoichiometric nature of many phosphosites, failure to accurately detect and quantify them can delay IND submissions, complicate tech transfers, or trigger costly re-analysis.
Phosphorylation analysis is now essential for antibody and protein-drug developers, offering insights that reduce development risk, support regulatory submissions, and remain fully RUO-compliant.
Phosphorylation Analysis with Creative Proteomics
Creative Proteomics offers high-sensitivity, high-accuracy protein phosphorylation analysis using state-of-the-art mass spectrometry. Whether you're optimizing a monoclonal antibody production process or exploring kinase inhibitors, our phosphorylation profiling platform delivers the insights needed to advance your project with confidence by identifying critical phosphosites and improving product comparability.
Our Services Include (but Are Not Limited To):
- Map phosphorylation dynamics under different biological conditions
- Comprehensive phosphorylation profiling: global and site-specific
- Site-specific phosphorylation identification and quantification
- Phosphopeptide enrichment strategies (TiO₂, IMAC, anti-pTyr)
- Protein–protein interaction analysis
Key Advantages of Our Phosphorylation Protein Analysis Service
High Sensitivity for Low-Abundance Phosphoproteins Detect and quantify low-mass phosphorylated peptides often missed by conventional methods.
Rapid Phosphorylation Mapping Streamlined workflow supports fast and reproducible site identification, ideal for time-sensitive studies.
Exceptional Accuracy and Reproducibility High-confidence phosphorylation assignments with consistent CVs <10% across replicates.
Automated Chromatographic Systems Automated LC enhances throughput, reduces manual error, and improves data consistency.
Experienced Scientific Team End-to-end support from experimental design to data interpretation.
Customizable Solutions Every project is tailored to your protein type, matrix complexity, and research goals.
The Science Behind Phosphorylation & Phosphorylation Sites
1. How a Phosphate Group Gets There
Kinases catalyze the transfer of the γ-phosphate from ATP to the hydroxyl groups of Ser, Thr, or Tyr residues. Phosphatases reverse the process. This dynamic modification modulates charge (adding ~–2 per phosphate), conformation, and molecular interactions.
Only a subset of the ~500 human kinases are active in CHO or HEK293 cells, explaining why recombinant biologics often show narrow phosphorylation profiles.
2. Hotspot Residues in Biologic Formats
Phosphorylation frequently localizes to:
- Hinge and CH2 regions in antibodies
- Linkers in Fc-fusions
- C-terminal tags in nanobodies
These regions are flexible and exposed, allowing kinase access. Modifications here can impact:
- pI shifts and charge-variant profiles (CEX)
- Thermal stability (Tm), via altered hydrogen bonding or repulsion
- FcγR/FcRn binding for CH2-adjacent phosphosites
4. Crosstalk with Other PTMs
| PTM Pair | Impact |
|---|---|
| Phospho ↔ Glyco | Thr near Asn297 reduces glycan occupancy by ~20% |
| Phospho ↔ Oxidation | Nearby Met oxidation destabilizes phosphosite |
| Phospho ↔ Deamidation | Acidic pH accelerates Asn deamidation + phospho-transfer |
Understanding these interactions helps upstream engineers and QC analysts reduce unwanted variability through pH control, kinase inhibition, or trace metal management.
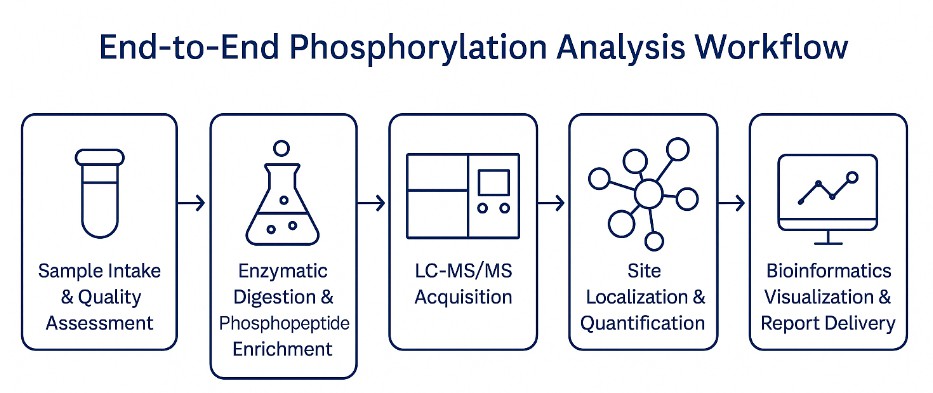
End-to-End Phosphorylation Analysis Workflow
At Creative Proteomics, our workflow integrates industry best practices with proprietary optimizations tailored to the challenges of analyzing phosphorylation in biopharmaceutical proteins.
Accepted Sample Types
| Sample Type | Typical Use Case | Requirements |
|---|---|---|
| Purified monoclonal antibodies (mAbs) | Comparability, lot-release analysis, charge-variant investigation | ≥ 50 µg, > 0.5 mg/mL, free from detergents and phosphate buffers |
| Fc-fusion proteins | Process development, stability studies, FcγR binding correlation | ≥ 100 µg recommended due to additional domain complexity |
| Recombinant proteins (cytoplasmic/secreted) | Kinase-substrate validation, PTM site discovery | > 70% purity preferred; include expression vector if possible |
| Cell culture supernatants | Early-stage clone screening, secreted phosphoprotein capture | ≥ 1 mL, filtered, with total protein > 100 µg |
| Protein digests or enriched phosphopeptides | For clients with in-house sample prep capacity | Must include digestion/enrichment protocol and peptide amount |
Phosphopeptide Enrichment Techniques
| Enrichment Method | Use Case | Strength |
|---|---|---|
| TiO₂ chromatography | Global phosphoproteome mapping | High recovery for mono- and multi-phosphorylated Ser/Thr peptides |
| Fe³⁺-IMAC columns | Orthogonal to TiO₂ for acidic sites | Selective for acidic phosphopeptides, reduces background |
| Anti-phosphotyrosine immunoprecipitation (IP) | Specific pTyr enrichment | >100-fold enrichment for low-abundance pTyr motifs |
LC-MS/MS Acquisition
High-resolution analysis is performed using Thermo Q Exactive HF-X or Exploris 480 Orbitrap systems with nano-UHPLC for maximum sensitivity. Acquisition methods include:
- DDA for site discovery
- DIA/PRM for targeted quantification of known sites
Typical benchmarks:
- LOD: 50–100 fmol per phosphopeptide
- CV: <10% across technical replicates
- Mass accuracy: ≤5 ppm
Site Localization & Quantification
We use MaxQuant, Byonic, and custom algorithms to:
- Assign site localization probabilities (≥0.75 = high confidence)
- Estimate relative site occupancy across conditions or lots
- Flag ambiguous sites and suggest validation strategies
- Results are cross-referenced with known domains (e.g., CH2, hinge, Fab) and literature annotations for interpretability.
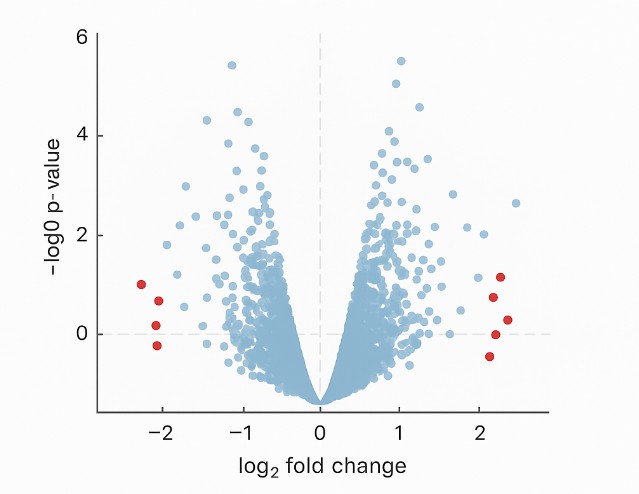 Volcano plot illustrating the distribution of phosphorylation sites based on log₂ fold change and –log₁₀ p-value. Significantly regulated sites are highlighted, facilitating the identification of differential phosphorylation across conditions.
Volcano plot illustrating the distribution of phosphorylation sites based on log₂ fold change and –log₁₀ p-value. Significantly regulated sites are highlighted, facilitating the identification of differential phosphorylation across conditions.Bioinformatics Visualization & Report Delivery
Clients receive:
- Annotated phosphopeptide lists with site-level confidence
- Volcano plots, heatmaps, and occupancy trend charts
- Overlay with charge variants or glycosylation if applicable
- Full raw MS data + processed files (e.g., Skyline, MaxQuant)
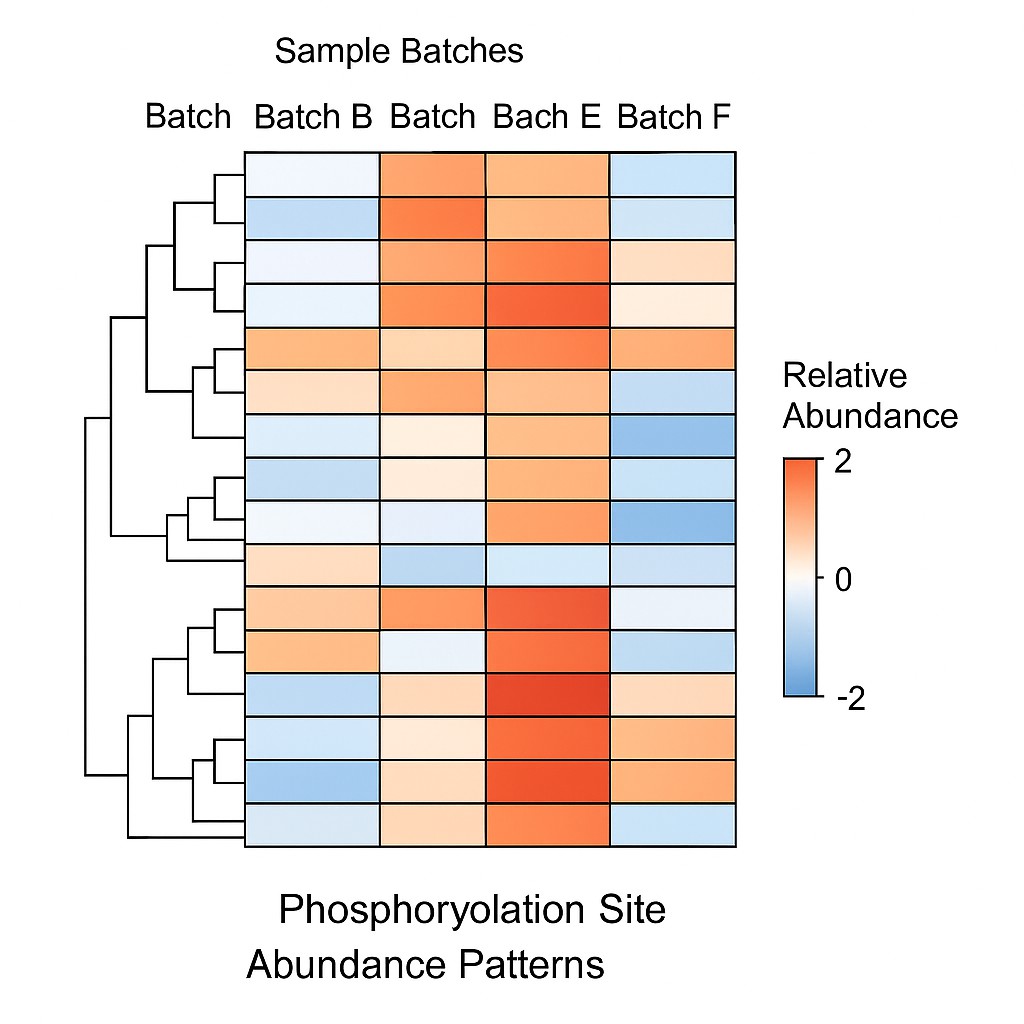 Heatmap showing the relative abundance of phosphorylation sites across different sample batches. The color gradient indicates site-specific expression variation and clustering.
Heatmap showing the relative abundance of phosphorylation sites across different sample batches. The color gradient indicates site-specific expression variation and clustering.Technical Highlights
| Metric | Platform Capability |
|---|---|
| Limit of Detection | 50–100 fmol |
| Localization Score Threshold | ≥0.75 (site-specific confidence) |
| Technical Reproducibility | CV <10% |
| Data Throughput | Up to 10 samples/week (discovery mode) |
| Enrichment Depth | TiO₂ + IP yields broad phospho coverage |
Applications of Phosphorylation Profiling
- Monitoring lot-to-lot consistency
- FcγR or FcRn binding correlation analysis
- Stability studies under stress (pH, temp, oxidants)
- Clone or expression system comparison
- Detecting process-induced PTM drift
Frequently Asked Questions (FAQ)
Q: Can we use phosphate-buffered samples?
A: Phosphate interferes with enrichment. We recommend buffer exchange or desalting. Our team can advise on preparation.
Q: Can you reanalyze for other PTMs later?
A: es. All raw files are archived for potential reprocessing (e.g., glycosylation, oxidation).
Q: Is data compatible with regulatory submissions?
A: While RUO only, our reports follow ICH Q6B and USP <1047> best practices and include detailed, reproducible metadata.
Q: Can we request only pTyr or only global phosphoproteome?
A: Yes. You may select site-specific enrichment, targeted PRM, or full profiling.
Q: What if we're unsure whether phosphorylation is relevant?
A: Our scientists offer no-obligation consults to help assess whether phosphorylation data could impact your product's profile.
Case Study: Phosphorylation Near Drug Binding Sites Affects Drug Efficacy
Reference
- Creixell, P., Palmeri, A., Miller, C. J., Lou, H. J., Santini, C. C., Nielsen, M., ... & Linding, R. (2015). Survey of phosphorylation near drug binding sites in the Protein Data Bank. Nature Communications, 6, 8641. DOI: 10.1002/prot.24605
Overview:
This study conducted a comprehensive analysis of phosphorylation sites in proximity to drug binding regions across 453 mammalian protein structures available in the Protein Data Bank (PDB). The aim was to assess how phosphorylation events near these sites might influence drug binding and efficacy.
Key Findings:
Prevalence of Phosphorylation Near Drug Binding Sites:
Approximately 29% of the analyzed drug target proteins had phosphorylation sites within 12 Å of the drug binding site. This proximity suggests a significant potential for phosphorylation to affect drug binding affinity.
Functional Impact on Drug Binding:
Phosphorylation near the binding site can either hinder or enhance drug binding. For instance, phosphorylation-induced conformational changes may obstruct the binding pocket or alter its electrostatic properties, thereby affecting the interaction with the drug molecule.
Implications for Drug Development:
The study highlights the necessity of considering phosphorylation states during drug design and development. Neglecting these modifications could lead to inaccurate assessments of drug efficacy and safety.
This case study underscores the importance of detailed phosphorylation analysis in the context of drug development. Creative Proteomics' Phosphorylated Protein Analysis Service is well-equipped to identify and quantify phosphorylation events, particularly those near critical functional domains such as drug binding sites. By leveraging advanced mass spectrometry techniques, the service can provide insights into how phosphorylation may influence drug-protein interactions, aiding in the optimization of therapeutic candidates.
Incorporating such case studies into your service page can effectively demonstrate the practical applications and value of phosphorylation analysis in biopharmaceutical research, thereby enhancing client engagement and conversion.
References
- Dephoure N., Gould K.L., Gygi S.P., et al. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Molecular Biology of the Cell. Volume 24 March 1, 2013. DOI: 10.1091/mbc.E12-09-0677
- Swaney, D., Beltrao, P., Starita, L. et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 10, 676–682 (2013). https://doi.org/10.1038/nmeth.2519
- McLachlin D.T. and Chait B.T. Analysis of phosphorylated proteins and peptides by mass spectrometry. Current Opinion in Chemical Biology, 2001, 5:591–602. DOI: 10.1016/s1367-5931(00)00250-7
- Han Y, Zhong L, Ren F (2022) A simple method for the preparation of positive samples to preliminarily determine the quality of phosphorylation-specific antibody. PLoS ONE 17(7): e0272138. https://doi.org/10.1371/journal.pone.0272138
Related Services
Support Documents