Peptide analysis is a main method to identify protein drugs and preparations. Peptide mapping can be used to decompose proteins by protease or chemical substances. Proper analytical methods can be used to identify the integrity and accuracy of primary structure of proteins, qualitative and quantitative analysis of protein degradation products. Peptide mapping has been used in many recombinant drugs as an important part of the primary structure confirmation and quality control.
We Provide but Are Not Limited to:
- Identification of primary structure or protein
- Determination of the complete sequence of amino acids (including its N-terminal and C-terminal)
- Identify the sites and proportions of protein-modified groups such as glycosylation, acetylation, sulfation, and phosphorylation
- Qualitative and quantitative analysis of degradation products of related proteins
- Identify the information of protein oxidation and deamination
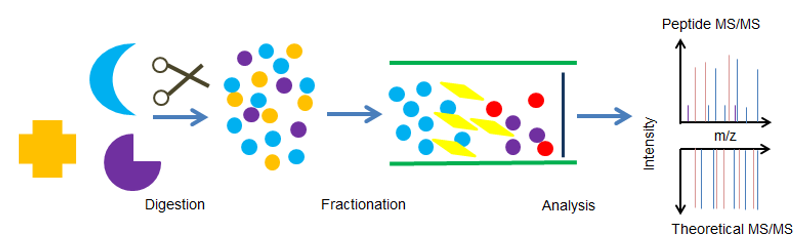
The Process of Peptide Mapping Service
- Six proteases (Trypsin, Chymotrypsin, ASP-N, Glu-C, LYS-C and LYS-N) are used for enzyme digestion and identification of target proteins
- Analyze samples by nano LC-MS/MS or UV LC-MS/MS
- Compare the complete sequence of the obtained peptide spectra with the theoretical sequence you provided
- Obtain and confirm the comparison results
Advantages of Peptide Mapping Service:
- The peptide map is based on the LC-MS / MS (series quality spectrum) technique. It can be combined with the combination of spectral detection and mass spectrometry. Compared with the LC-MS or MALDI-TOF basis, our peptide spectrometry can give accurate conclusions to the first level structure and the modification group of the protein.
- Our peptide map analysis can give accurate information about the small changes in the concentration and the incomplete separation of the liquid chromatography.
- The software tools of the ProtTechPepMap can fully retrieve various possible modifications of the report in the literature, and can also retrieve the embellifiers specified by the customer.
Sample Requirements
- The in-gel sample or solution sample can be used for the above sequencing
- The purity of the sample is not less than 80% protein
- Required target protein sample size: 10-20 µg
- Protein glue samples may also be used for the determination of peptide coverage, but special pretreatment is required
Creative Proteomics' analytical scientists can provide a clear and concise peptide atlas analysis report to help clients detect protein level structure, amino acid sequence, etc.
Case Analysis of Peptide Mapping Service:
In accordance with the principle of improving efficiency and reducing the cost of the customer, the analysis process is based on the prediction of the peptide segment coverage according to several different protease enzymes, and if you can only cover the whole sequence by three kinds of egg white enzyme, the actual experiment will use these three egg white enzymes to analyze the sequence of enzymes and subsequent sequences, which can effectively reduce the cost of the experiment. The following is part of the previous project, which is shown in the theoretical sequence of antibody drugs, and the first order sequence needs to be validated to demonstrate that the antibodies are consistent with the theoretical sequence, and some experimental data are shown as follows:
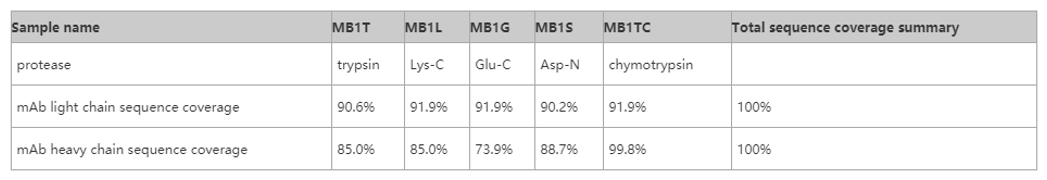
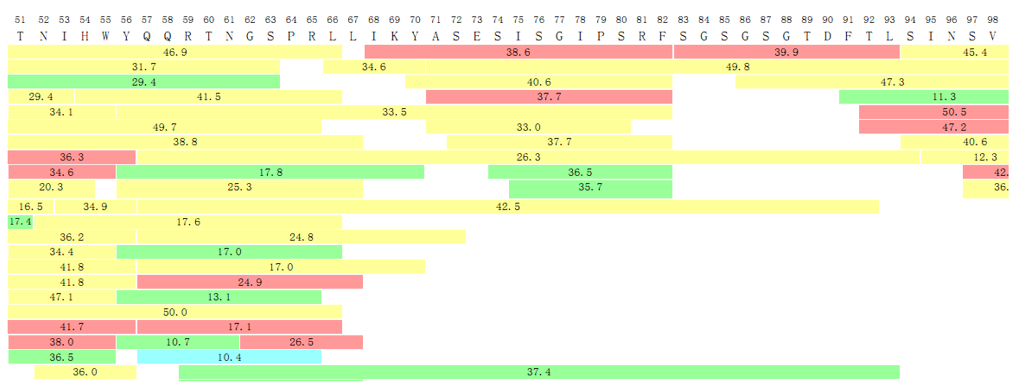
In the final report, we will give detailed data on the coverage of the peptide section as shown in the following diagram.
References
- Wen-Shan Z, Hai-Ming L, et al. Significance of Peptide Mapping in the Quality Control of Recombinant Human Parathyriod Hormone 1-34 Product. chinese pharmaceutical journal, 2006.
- RAO C M, ZHANG Y, HAN C M, et al. Peptide mapping analysis of Rewnbinant human interleuldn-11 by tryptic digestion. Acta Pharm Sin, 2000, 35(5): 378-380.






