Protein molecules are biological macromolecules formed by many amino acids connected by peptide bonds. Physiologically functional proteins in the body are all ordered structures. Each protein has a certain percentage of amino acid mass, the sequence of amino acids, and the specific arrangement position of the peptide chain space. Therefore, the protein molecular structure composed of the sequence of amino acids and the spatial arrangement of peptide chains is the structural basis for each protein to have a unique physiological function.
There are 20 kinds of amino acids that make up human protein, and the molecular weight of proteins is relatively large. The sequence and spatial position of amino acids in proteins are almost endless. Different amino acid sequences and specific spatial arrangements can create tens of thousands of proteins in the human body, and complete tens of millions of physiological functions endowed by life.
In 1952, Danish scientists suggested dividing the complex molecular structure of proteins into 4 levels, namely primary, secondary, tertiary, and quaternary structures. The latter three are collectively referred to as higher-level structures or spatial conformations.
In protein molecules, the sequence of amino acids from N-terminus to C-terminus is called the primary structure of the protein. The main chemical bonds in the primary structure are peptide bonds. In addition, the positions of all disulfide bonds in protein molecules also belong to the category of the primary structure. It is a one-dimensional structure without a spatial concept, and with the deepening of research, it is realized that a protein's primary structure is not the only factor that determines the spatial conformation of a protein.
There are many complex proteins in the living body. Besides amino acids and peptide chains, proteins also have other components, such as sugar chains on glycoproteins, lipid parts in lipoproteins, etc.
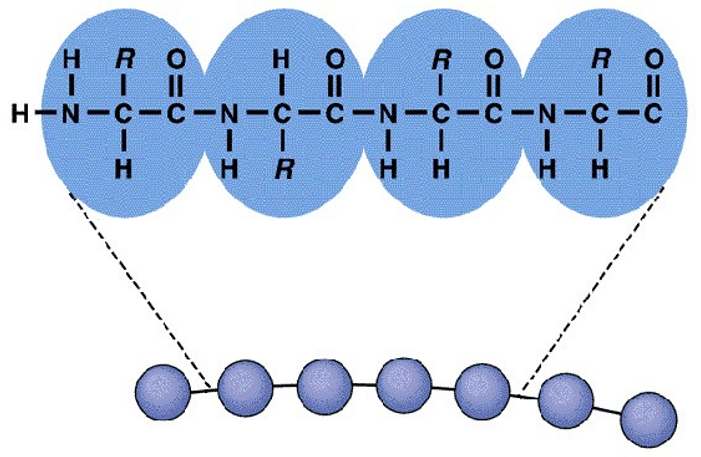
Protein primary structure characterization
To characterize the protein primary structure, common methods are:
- Amino acid sequencing
- Edman N-terminal sequencing
- C-terminal sequencing
- Molecular weight determination
- Qualitative and quantitative determination of modifications (such as oxidation, deamination, acetylation, methylation and crosslinking)
1. Edman degradation N-terminal sequencing
Edman degradation sequencing can label and analyze the N-terminal amino acid sequence without disturbing the peptide bond. But this method is not suitable for N-terminal blocking or chemical modification.
2. Protein sequence analysis based on mass spectrometry
In the process of protein sequence analysis, six proteases (trypsin, chymotrypsin, Asp-N, Gluc-C, Lys-C, and Lys-N) are usually used to digest the target protein, reliably isolate and identify the resulting peptide, and then gain insight into the complete sequence information of the protein. While the peptide fragments are obtained by mass spectrometry, 100% determination of the protein sequence is completed through the splicing between the peptides. Each amino acid composition and surrounding amino acid microenvironment can then be displayed, including disulfide bond information. Structural characterization performed at this level highlights post-transcriptional modifications (PTM) such as site-specific glycosylation, amino acid substitutions (sequence variants) and/or truncations resulting from incorrect transcription of complementary DNA.






