Venom Transcriptome Assembly
Venom transcription analysis is an important tool for venom genome, biological activity, toxicological and pharmacological research. Simply comparing the reads with a reference genome by transcriptome sequencing can give a rough estimate of venom gene expression, but if you want to figure out the exact performance of venom gene activity, you must assemble a set of reads into a transcript. Creative Proteomics uses standardized experimental procedures, advanced sequencing platforms, and professional information analysis to provide high-efficiency and high-quality venom transcriptome assembly services for venom research and a basis for subsequent venom gene expression and biological function research.
Why Choose Venom Transcriptome Assembly
In order to explore the toxicology and therapeutic pharmacology of venom damage in different toxic species, more transcriptome information about these species is needed to facilitate further research on target genes. The venom transcriptome assembly results will help us to study the venom genes and their biological activities for venomous species such as snakes, wasps, scorpions, etc. The venom transcriptome assembly mainly uses two methods: de novo assembly and reference guidance. De novo assembly is to find the overlap between the reads and try to link them together to form a complete transcript. Reference-guided assembly uses the existing genome, first splice the sequencing reads and compare, and then construct a splicing map based on the comparison, use these maps to construct individual transcripts.
Our Venom Transcriptome Assembly Services
There are still many venom studies using a single assembly method, such as snake venom metalloprotease which is difficult to assemble. Creative Proteomics provides solutions to these problems through the combination of several venom transcriptomics assembly strategies to complete the difficult assembly of venom transcriptome sequencing reads. Our methods include but are not limited to the following:
- Trinity. The common method that venom transcriptome assembly based on de Bruijn.
- SOAPdenovo-Trans. De novo transcriptome assembly for short RNA sequence reads.
- BinPacker. Packaging-based de Novo transcriptome assembly from RNA sequence data.
- Oases. Robust de novo RNA sequence assembly within the dynamic range of expression levels.
- VTBuilder. A tool for assembling multiple subtype transcriptomes.
Data Analysis
Creative Proteomics provides customers with the analysis of the results of venom assembled transcriptomes and evaluates the integrity of the toxin gene assembly in the venom and the quality indicators of the venom transcriptome assembly.
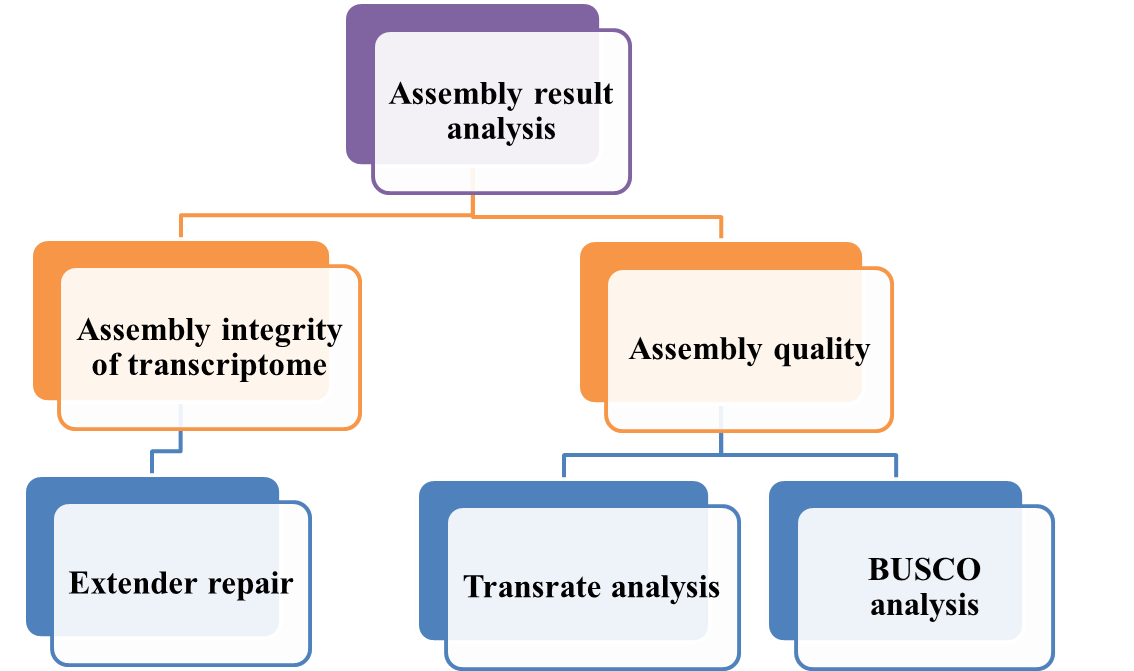
- A single assembly method cannot assemble the most complete venom transcriptome. We select complementary venom transcriptome assembly programs based on the results of transcription integrity assessment to achieve the most complete venom transcriptome.
- We use two common assembly quality indicators to analyze the venom transcriptome assembly results. BUSCO recognizes single copy, orthologous non-toxin loci in each assembly, and classifies the annotated contigs as complete and single copies, complete and duplicates, fragmented or missing. Transrate assesses contigs for base calling and structural chimerism or other forms of misassembly.
Our Advantages
- We combine and screen the venom transcriptome assembly results using various assembly strategies to obtain complete high-quality sequences.
- The evaluation of the assembly results of venom transcriptomics is conducive to follow-up research.
- Usable methods and practical suggestions for the application of venom transcriptome assembly in the study of venom gene evolution.
With the rise of transcriptomics research, available methods for venom transcriptome assembly are also emerging in endlessly. Creative Proteomics provides a variety of venom transcriptome assemblers and binding strategies to change the result deviation caused by a single assembly method in the current venom research and provide customers with a highly complete and high-quality venom transcriptome assembly result.
If you are interested in our services, please feel free to contact us.
Reference
- Matthew H, et al. Evaluating the Performance of De Novo Assembly Methods for Venom-Gland Transcriptomics. Toxins, 2018, 10(6):249.
For research use only. Not intended for any clinical use.

