Venom Epigenome Sequencing
Creative Proteomics as one of the venomics research service providers, it provides different types of epigenome sequencing services for venom research with its rich epigenome sequencing experience, so as to meet the needs of epigenome sequencing for different venom research purposes. Creative Proteomics provides whole-genome bisulfite sequencing (WGBS), reduced-representation bisulfite sequencing (RRBS), MeRIP-Seq/m6A-seq, ChIP-seq, RIP-seq, and other levels of epigenome sequencing services for different poisonous species (such as snakes, scorpions, frogs, venomous lizards, fire-bellied toads, snails, spiders, bees, ants, cone snails, jellyfish, etc.) to meet the needs of various levels of scientific research.
Introduction

Epigenomes (genetic modifiers) effectively mark genomes epigenomes not to change the DNA sequence but to change the way cells use the DNA’s instructions. These marks (such as histone modification, chromatin Immuno-precipitation, etc. ) can pass to other cells and they can also be passed down through family lines. Epigenomes are tiny chemical compounds that attach to DNA, and they play a crucial role in regulating gene activity. In venom research, epigenomic modifications, for example, may shut off or turn on a gene or control protein production in particular cells. Ultimately regulate the expression and synthesis of venom-related components.
What Creative Proteomics Offers
Creative Proteomics has rich experience in epigenome sequencing projects and poisonous species experience, and the different type of epigenome library constructed by it has a good success rate and stability. In addition, the sequencing quality, genome comparison rate, coverage rate and other quality control indicators are also excellent. In venom research, Creative Proteomics provides different type of epigenome sequencing for researchers.
- Whole-genome bisulfite sequencing (WGBS): DNA methylation is an important epigenetic marker information. Obtaining the data of all methylation sites in the whole genome is of great significance for the study of temporal and spatial specificity of venom epigenetics.
- Reduced-representation bisulfite sequencing (RRBS):RRBS uses restriction enzymes to digest the genome, and then conducts Bisulfite sequencing to analyze the methylation of the genome. In venom research, this technology has significantly improved the sequencing depth of CpG regions, and high-precision resolution can be obtained in CpG islands, promoter regions and enhancer element regions.
- MeRIP-Seq/m6A-seq: MeRIP-Seq maps m6A-methylated RNA. In this sequencing method, m6A-specific antibodies are used to immunoprecipitate RNA. RNA is reverse-transcribed to cDNA and sequenced. Deep sequencing provides high-resolution reads of m6A-methylated RNA.
- ChIP-seq: ChIP-seq is a powerful tool for studying protein-DNA interactions, and is usually used for the study of transcription factor binding sites or histone-specific modification sites. The ChIP-Seq technology, which combines ChIP and next-generation sequencing technology, can efficiently detect DNA segments that interact with histones and transcription factors in the whole genome in venom research.
- RIP-seq: RIP-seq is a technology of RNA immunoprecipitation combined with high-throughput sequencing, which captures the interacting RNA by immunoprecipitating the target protein. The combination of RIP technology and high-throughput sequencing can help researchers understand the dynamic process of post-transcriptional regulatory networks at the whole genome level.
Epigenome Sequencing Workflow
The venom epigenome sequencing service provided by Creative Proteomics, from different sample processing (such as chromatin Immunoprecipitation, RNA immunoprecipitation, etc.), library construction and to sequencing, has undergone scientific and meticulous design and strict quality control at every step to ensure high-quality sequencing results. The following figure shows the general process of reduced-representation bisulfite sequencing:
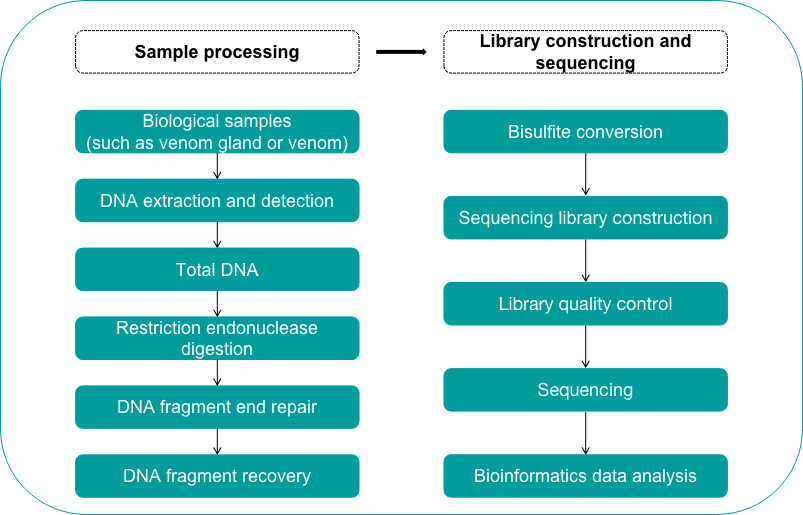 Fig 1. Venom reduced-representation bisulfite sequencing workflow - Creative Proteomics
Fig 1. Venom reduced-representation bisulfite sequencing workflow - Creative Proteomics
Creative Proteomics, as a rare professional venomics research service provider in the world, relies on its years of sequencing experience to provide customers with one-stop venom epigenome sequencing services. If you have any questions about our venom epigenome sequencing service, please feel free to contact our technical support, the technical support will reply to you as soon as possible!
References
- Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons[J]. Cell. 2012 Jun 22;149(7):1635-46.
- Gokhale NS, et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection[J]. Cell Host Microbe. 2016 Nov 9;20(5):654-665.
For research use only. Not intended for any clinical use.


 Fig 1. Venom reduced-representation bisulfite sequencing workflow - Creative Proteomics
Fig 1. Venom reduced-representation bisulfite sequencing workflow - Creative Proteomics