Why Food Peptidomics?
Food peptidomics provides a direct, high-resolution view of endogenous and process-generated peptides in complex matrices. Unlike protein-only or amino-acid assays, peptidomics explains what peptides are present, at what levels, and how processing changes them. This evidence enables:
- Claim support: Quantify ACE-inhibitory, antioxidant, satiation-related, and mineral-binding peptides.
- Process optimization: Track hydrolysis, fermentation, thermal steps, and Maillard-related changes at peptide resolution.
- Allergen risk management: Detect epitope-containing peptides and digestion-resistant fragments.
- Quality differentiation: Compare lots, suppliers, and batches with quantitative peptide fingerprints.
What Makes Food Peptidomics Powerful?
Food peptidomics delivers molecular-level insights that protein or amino-acid tests miss. It links peptide identity, quantity, and modification to process and performance.
- Depth beyond proteins: Reveals functional fragments from enzymes, fermentation, and digestion.
- Quantitative precision: Targeted MRM/PRM enables absolute or relative peptide levels.
- Process traceability: Captures thermal, enzymatic, and Maillard fingerprints across batches.
- PTM visibility: Detects glycation, oxidation, deamidation, and cross-links that alter behavior.
- Bioactivity annotation: Maps ACE-inhibitory, antioxidant, calcium-binding, or satiation motifs.
Key Application Areas
Food peptidomics bridges the gap between protein hydrolysis, processing, and functional activity—quantifying peptides that shape flavor, bioactivity, and stability across the food value chain.
What We Offer: Comprehensive Food Peptidomics
Our food peptidomics platform transforms complex peptide mixtures into interpretable, quantitative datasets. Whether you're screening unknown bioactive peptides or validating known motifs, we provide data you can act on with confidence.



ACE-inhibitory, DPP-IV, antioxidant, calcium-binding, appetite/satiety-related motifs.

Types of Peptides Detectable by Food Peptidomics
| Peptide Type | Source / Generation Mechanism | Scientific Relevance |
|---|---|---|
| Endogenous peptides | Naturally present in raw matrices (e.g., dairy, soy, meat) | Baseline nutritional content; precursor for bioactive motifs |
| Enzymatic hydrolysate peptides | Produced by proteases such as trypsin, alcalase, flavourzyme | Functional ingredient screening; bitterness vs bioactivity balance |
| Fermentation-derived peptides | Microbial proteolysis during fermentation | Strain-specific profiles; quality fingerprinting |
| Digestion-resistant peptides | Survive in-vitro gastric/intestinal digestion | Epitope persistence; in-vivo stability modeling (research-only) |
| Allergenic/epitope-bearing peptides | Peptides containing known IgE-binding motifs (research-only) | Risk screening in processed food, R&D allergen studies |
| Process-modified peptides | Altered by heat, oxidation, pressure, glycation | Indicators of shelf-life, degradation, or Maillard reactions |
| Post-translationally modified peptides (PTMs) | Oxidation, deamidation, cross-linking, glycation | Functional impact on structure, bioactivity, and flavor |
| Bioactive peptide motifs | Known functional sequences (e.g., ACE-I, DPP-IV inhibitors) | Health-related claims; functional product formulation |
Notes:
- Quantitation of these peptides is matrix- and sequence-dependent. We use DIA/DDA for discovery and MRM/PRM for targeted quantification.
- Peptides are annotated against public databases and motif libraries where applicable (e.g., BIOPEP-UWM, IEDB, MilkAMP).
Platform Advantages
High-Sensitivity Peptide Detection

Comprehensive Modification Profiling

Matrix-Tolerant Sample Handling

Flexible Quantification Modes

Accurate Peptide-Level Fingerprinting

Low Sample Input Compatibility
Step-by-Step Food Peptidomics Workflow
- Discovery mode: Data-Dependent (DDA) or Data-Independent Acquisition (DIA/DIA-NN) with 30–120 min C18 gradients.
- Targeted mode: Scheduled PRM/MRM with isotope-labeled standards using Orbitrap or triple quadrupole platforms.
High-Resolution Instrumentation for Food Peptidomics
At Creative Proteomics, our food peptidomics workflow is built on cutting-edge mass spectrometry platforms optimized for short, modified, and low-abundance food-derived peptides. From discovery profiling to targeted quantification, we deliver deep peptidome coverage with high reproducibility across complex matrices such as hydrolysates, fermented foods, and plant/animal extracts.
Our integrated platform combines Orbitrap Exploris™, timsTOF Pro (diaPASEF), and triple quadrupole systems to meet the analytical demands of both research and industrial peptide analysis.
Technical Highlights
- High-Resolution Orbitrap MS
Full MS at up to 120k resolution with ≤3 ppm mass accuracy using lock-mass calibration. - Ion Mobility Separation (timsTOF)
diaPASEF acquisition with >100 Hz MS/MS rate and CCS filtering for dense food matrices. - Flexible Acquisition Modes
Supports DDA/DIA for discovery and PRM/MRM for targeted quantification. - Low Detection Limits
Achieves ng/mL-level LOQs with internal standards and optimized extraction; CVs typically ≤10%. - Wide Dynamic Range
105 (discovery) and >106 (targeted), suitable for major and trace-level peptides. - PTM and Processing Modification Detection
Confident identification of glycation, oxidation, deamidation, and cross-links without enrichment. - Integrated QC System
Includes iRT calibration, blank/carryover checks, and pooled QC with drift correction.
Instrument Capability Overview
| Feature | Orbitrap Exploris™ / HF-X | timsTOF Pro (diaPASEF) | Triple Quadrupole (6500+) | Orbitrap PRM Mode |
|---|---|---|---|---|
| Scan Speed | ~40 Hz | >100 Hz | N/A (point-to-point) | ~10–20 Hz |
| MS/MS Coverage | >90% | >90% | Targeted only | High |
| PTM Sensitivity | High (glycation, oxidation, etc.) | High + CCS | Moderate (only if targeted) | High |
| Quantification Mode | Label-free, DIA | Label-free, DIA | MRM with isotope standards | PRM with accurate mass |
| LOQ Sensitivity | ~low ng/mL (discovery) | ~low ng/mL (DIA) | ng/mL to sub-ng/mL | ~low ng/mL |
| Dynamic Range | >105 | >105 | >106 | >105 |
| Best Use Case | Untargeted discovery, PTM mapping | High-throughput DIA profiling | Absolute quantification of known peptides | High-specificity quant of validated targets |
| Sample Input | 1–5 g solids / 20–50 mL liquid | Same | Matrix matched, ~1–10 mL or extract | Same |
Sample Requirements for Food-Derived Peptide Profiling
| Matrix Type | Minimum Amount* | Container | Preparation Notes | Storage & Shipping |
|---|---|---|---|---|
| Powders (protein isolates, hydrolysates) | 2–5 g | Sterile screw-cap tube | Provide enzyme program/process info if available. | Store at 2–8 °C dry; ship cold pack. |
| Liquids (milk, beverages, broths) | 50–100 mL | Sealed polypropylene bottle | Avoid thickeners/preservatives when possible. Record pH. | 2–8 °C; ship cold pack. |
| Fermented products | 50–100 g or mL | Sterile container | Record strain/process parameters; avoid live overgrowth. | 2–8 °C; ship cold pack; avoid freezing if carbonated. |
| Meat/Plant tissues | 50–100 g | Whirl-Pak®/cryovial | Trim bones/peels; note treatment (thermal/pressure). | -20 °C; ship on dry ice. |
| Oils/High-fat matrices | 20–50 mL | Amber vial | Provide antioxidant info; minimize headspace. | 2–8 °C; ship cold pack. |
| Gastrointestinal digests (in-vitro models) | 20–50 mL | Screw-cap tube | Clarify enzyme recipe and time points. | -20 °C; ship on dry ice. |
*If material is scarce, contact us for low-input options. Avoid antimicrobial preservatives. Document pH and key processing parameters for best results.
Demo Results
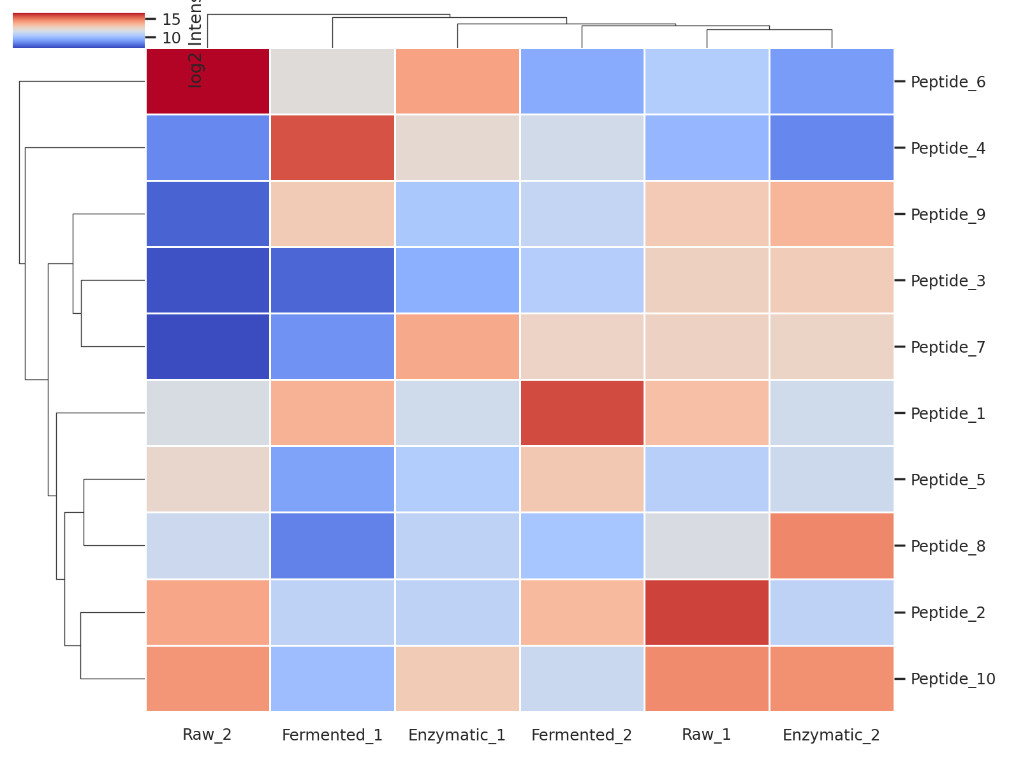
Peptide Intensity Heatmap with Hierarchical Clustering
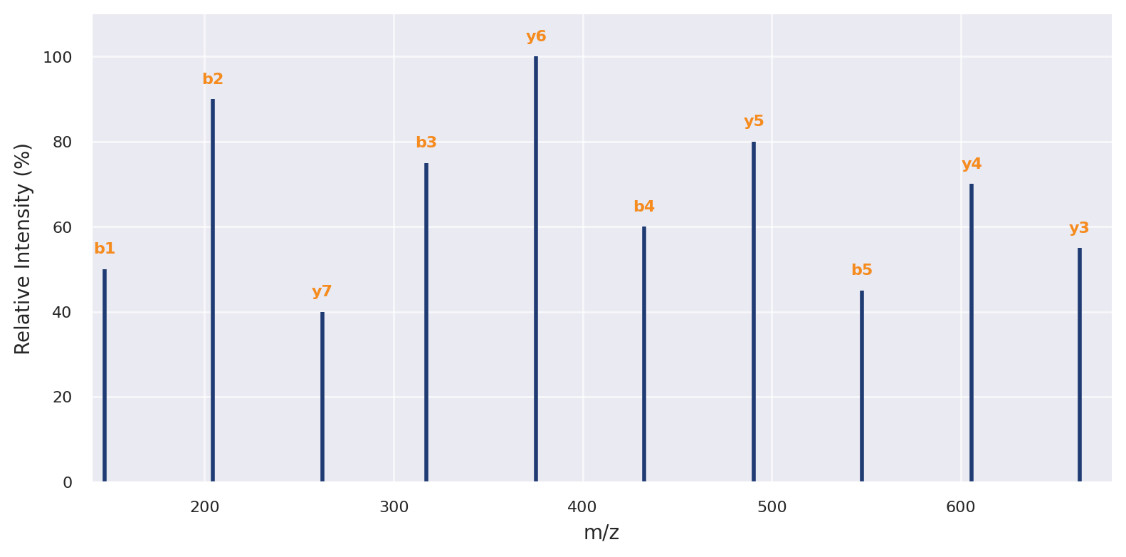
MS/MS Spectrum of a Representative Bioactive Peptide
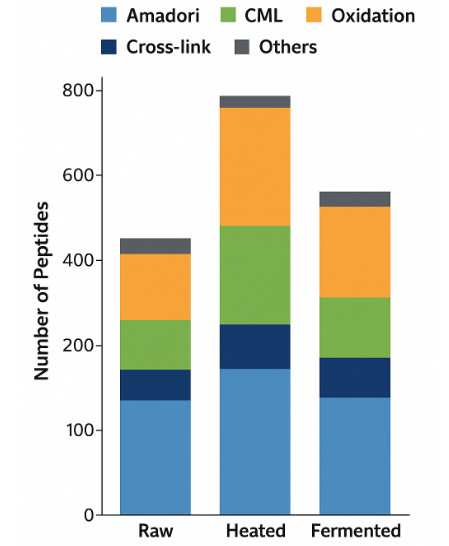
Peptide Modification Distribution Plot
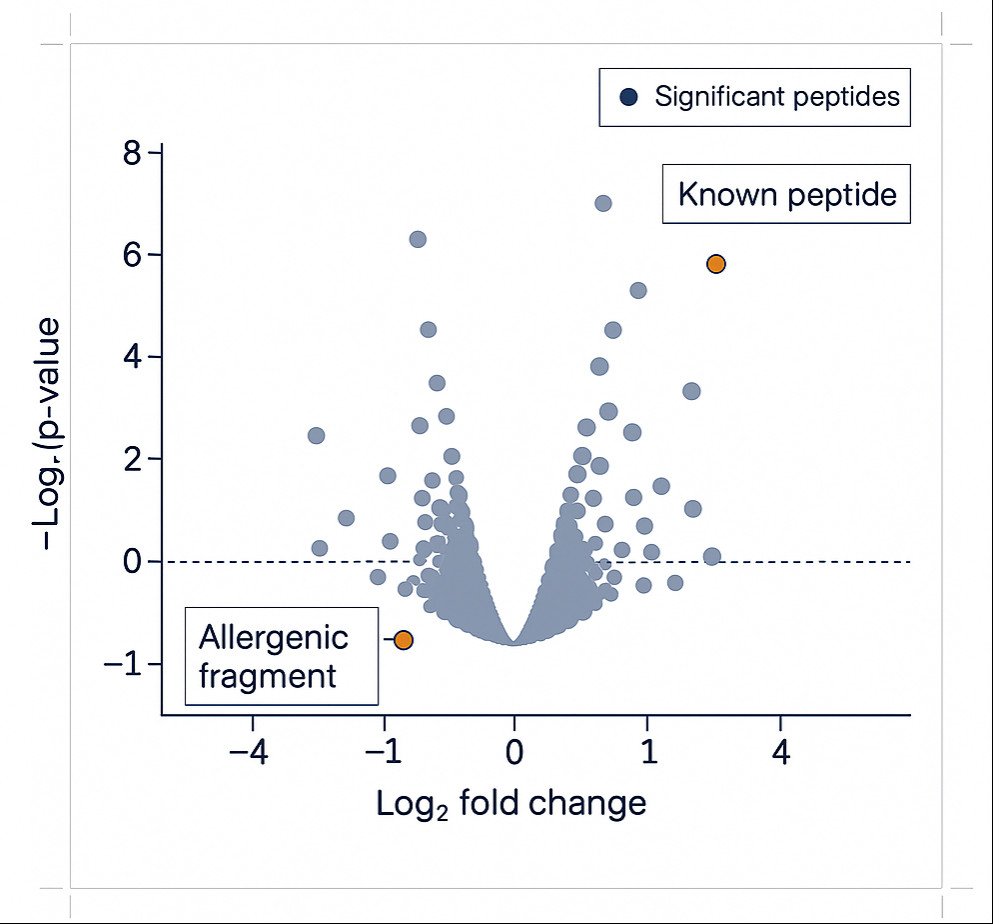
Volcano Plot for Differential Peptidome Profiling
Deliverables | What You Will Receive
- Raw & Peak Data
LC-MS/MS raw files (e.g., *.raw,.d) and processed peak lists (.mzML, *.mgf). - Peptide Identification
Annotated peptide lists with sequence, PTMs, precursor info, and confidence metrics (FDR/q-value). - Quantitative Results
Peptide intensity matrix across samples, differential analysis (fold change, p-value), CV%, and key statistical plots (e.g., PCA, volcano). - Precursor Protein Mapping
Links mature peptides back to their prohormone or precursor protein. - High-Quality Visuals
Publication-grade images: heatmaps, spectra, PTM distribution, and clustering plots (PNG/PDF/SVG formats). - Summary Report (PDF)
Method overview, QC stats, modification profiles, major findings, and optional biofunctional insights (e.g., via BIOPEP).

Combined Omics Approaches Reveal Resistance Mechanisms to Beet Curly Top Virus in Sugar Beet Genotypes
Journal: International Journal of Molecular Sciences
Published: 2023
https://doi.org/10.3390/ijms241915013BRCA1 Antibodies Matter: Benchmarking Reagent Specificity in Cancer Research
Journal: International Journal of Biological Sciences
Published: 2021
https://doi.org/10.7150/ijbs.63115In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1 and Effect of Phorbol Ester on Activation by TcPKC of TcPolβ in Trypanosoma cruzi Epimastigotes
Trypanosoma cruzi DNA Polymerase β Is Phosphorylated In Vivo and In Vitro by Protein Kinase C (PKC) and Casein Kinase 2 (CK2)
The Interplay Between GSK3β and Tau Ser262 Phosphorylation During the Progression of Tau Pathology
Journal: International Journal of Molecular Sciences
Published: 2022
https://doi.org/10.3390/ijms231911610Inactivation of Myeloperoxidase by Benzoic Acid Hydrazide
Journal: Archives of Biochemistry and Biophysics
Published: 2015
https://doi.org/10.1016/j.abb.2015.01.028





 Orbitrap Exploris™ 480
Orbitrap Exploris™ 480 Q Exactive HF-X
Q Exactive HF-X timsTOF Pro
timsTOF Pro Triple Quad™ 6500+
Triple Quad™ 6500+