Why Immunopeptidome Profiling?
The immunopeptidome—comprising peptides naturally presented by HLA molecules—serves as a real-time molecular signature of cellular state. It reveals changes driven by mutations, infections, or immune dysregulation long before phenotypic symptoms appear.
By profiling these HLA-bound peptides, researchers gain an actionable readout of the immune system’s interaction with tumor cells, pathogens, or self-antigens.
What Makes Immunopeptidomics Powerful?
Unlike transcriptomics or proteomics, immunopeptidomics directly captures what the immune system can "see"—the actual surface-displayed peptides recognized by T cells. It bridges the gap between expression and immune recognition.
Key Application Areas
When paired with genomics, transcriptomics, and proteomics, immunopeptidomics adds a functional layer that reveals how molecular changes translate into immune-detectable differences—crucial for drug discovery, biomarker validation, and precision immunotherapy.
What We Offer: Comprehensive Immunopeptidome Profiling and Beyond
Creative Proteomics provides an end-to-end immunopeptidomics service for the in-depth characterization of naturally presented HLA-bound peptides. By integrating advanced mass spectrometry platforms, custom enrichment workflows, and sophisticated bioinformatics pipelines, we empower researchers to identify, quantify, and interpret the dynamic antigen landscape in both physiological and disease states.
What We Can Analyze




Specialized Add-On Modules (Optional)



Platform Advantages
Low Sample Input Compatibility

Dual-Class HLA Analysis

HLA Binding Motif Characterization

Quantification-Ready Workflows

HLA Deconvolution & Allele Assignment

Comprehensive Bioinformatics Outputs
Step-by-Step Immunopeptidome Profiling Workflow
Deep and Accurate Peptidome Discovery
At Creative Proteomics, our immunopeptidomics workflow is powered by next-generation Orbitrap mass spectrometry platforms—Orbitrap Astral™ and Orbitrap Exploris™ 480—ensuring exceptional sensitivity, resolution, and throughput for comprehensive HLA ligand profiling.
Whether you're working with rare cell types or large cohorts, our platform delivers reproducible, deep-coverage datasets that accelerate epitope discovery and immunological insight.
Technical Highlights
- >90% MS/MS Coverage
High-resolution tandem MS enables identification of thousands of HLA-bound peptides with superior confidence and spectral clarity. - 1% FDR Stringent Filtering
Strict false discovery rate (FDR) control ensures reliable peptide identification and reproducibility across biological replicates. - PRM & SureQuant™-Enabled Quantification
Supports both relative and absolute quantification, with enhanced sensitivity for low-abundance immune peptides. - Broad Input Range: 1M–100M Cells
Optimized for scarce biospecimens or bulk materials—our platform scales with your project’s needs.
Orbitrap Astral™ vs. Orbitrap Exploris™ 480 — Technical Comparison
| Feature | Orbitrap Astral™ | Orbitrap Exploris™ 480 |
|---|---|---|
| MS/MS Coverage | >90% with high-speed, deep proteome coverage | >90% with robust, reproducible coverage |
| FDR Filtering | 1% FDR stringent filtering | 1% FDR stringent filtering |
| Quantification Support | PRM / SureQuant™ for absolute quantification | PRM / SureQuant™ for absolute quantification |
| Sample Input Range | Deep coverage from 100K–100M cell input | Deep coverage from 1M–100M cell input |
| Scan Speed | Up to 200 Hz (ultra-high speed) | ~40 Hz (high speed) |
| Sensitivity | Ultra-high sensitivity (single-cell compatible) | High sensitivity |
| Mass Accuracy | Sub-ppm mass accuracy (Orbitrap-based) | Sub-ppm mass accuracy (Orbitrap-based) |
Sample Requirements for Immunopeptidomes
| Sample Type | Minimum Amount | Preservation | Shipping Condition | Notes |
|---|---|---|---|---|
| Cell Pellet | ≥1×10⁶ cells | Snap-frozen (dry ice) | Dry ice, overnight courier | Avoid freeze-thaw; PBS wash preferred before freezing |
| Adherent Cells | ≥80% confluent in 6-well plate × 2 | Harvested & frozen as pellet | Dry ice, avoid liquid medium | Include cell count and viability if possible |
| Tissue (Fresh/Frozen) | ≥5 mg | Snap-frozen preferred | Dry ice | No fixatives or embedding media (e.g., paraffin) |
| PBMCs | ≥1×10⁶ viable cells | Cryopreserved in DMSO or fresh | Dry ice or room temp for fresh overnight | Prefer Ficoll-purified human PBMCs |
| Tumor Tissue | ≥10 mg | Snap-frozen or OCT embedded | Dry ice | Provide pathology report if available |
| FFPE Tissue (upon request) | ≥2 unstained 10 μm sections | FFPE block or slides | Ambient (slides) / Dry ice (block) | Please consult us prior to submission; yield may vary |
| Whole Blood | ≥500 μL (for PBMC prep) | EDTA tube, processed ≤2 h | Cold pack, same-day delivery | For downstream PBMC isolation; not ideal for direct MS |
Demo Results
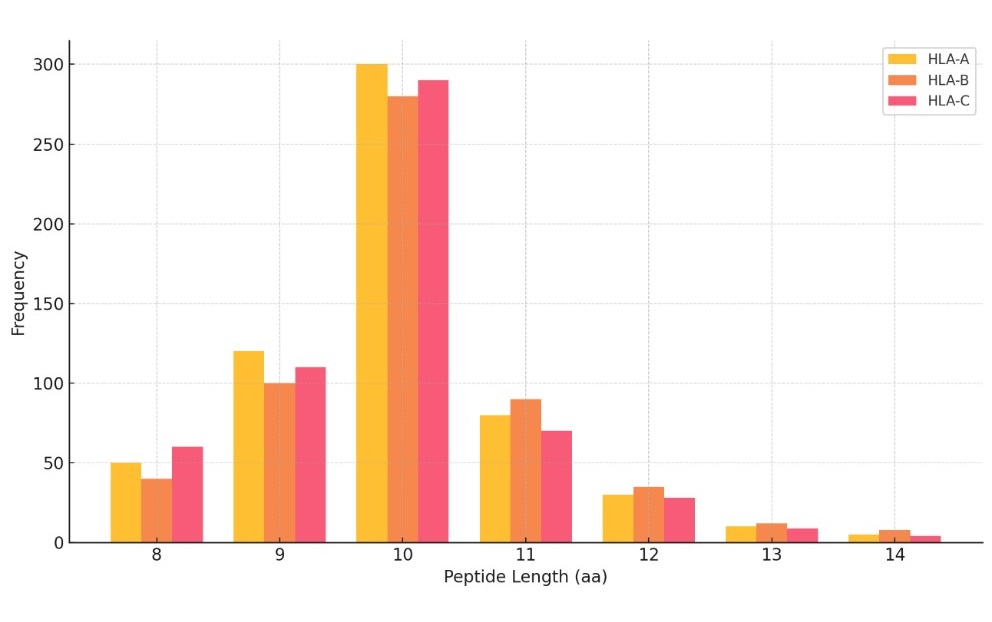
Peptide Length Distribution by HLA Type
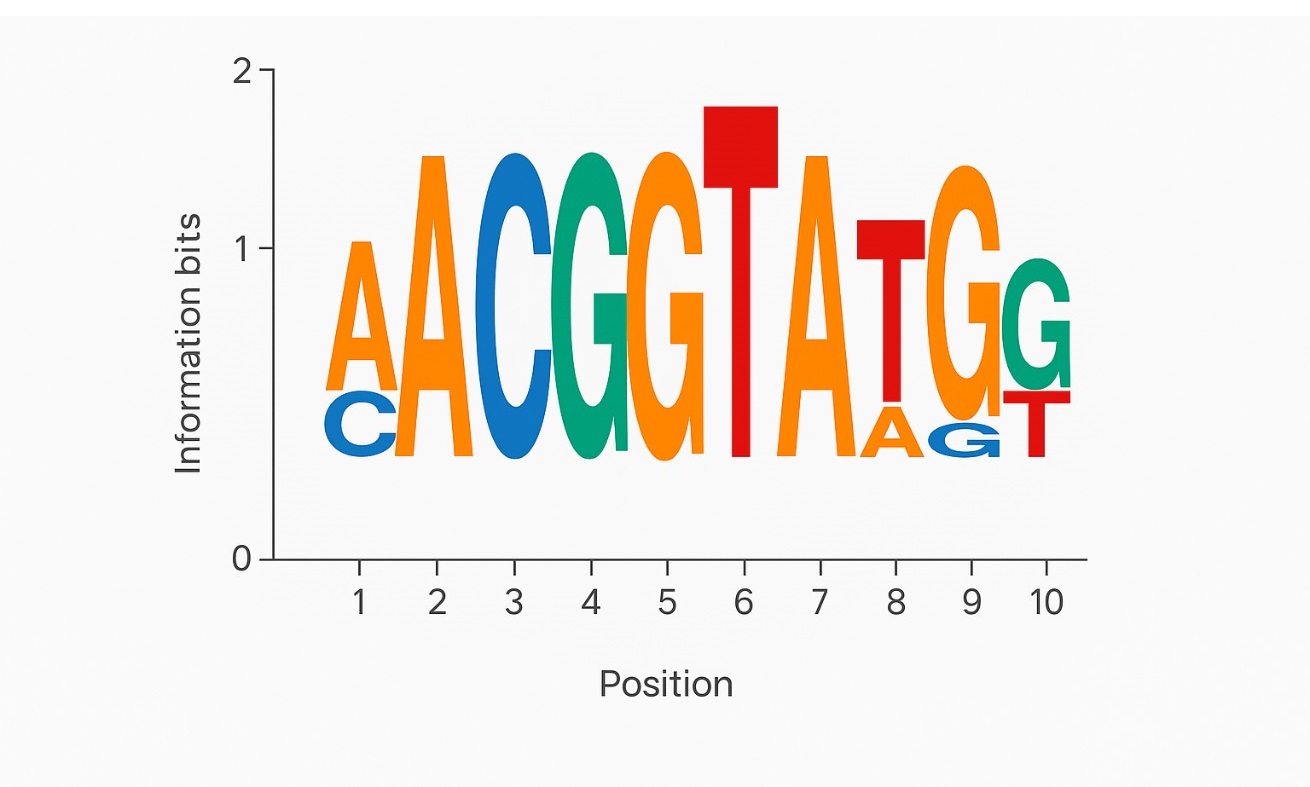
Motif Logo Plot
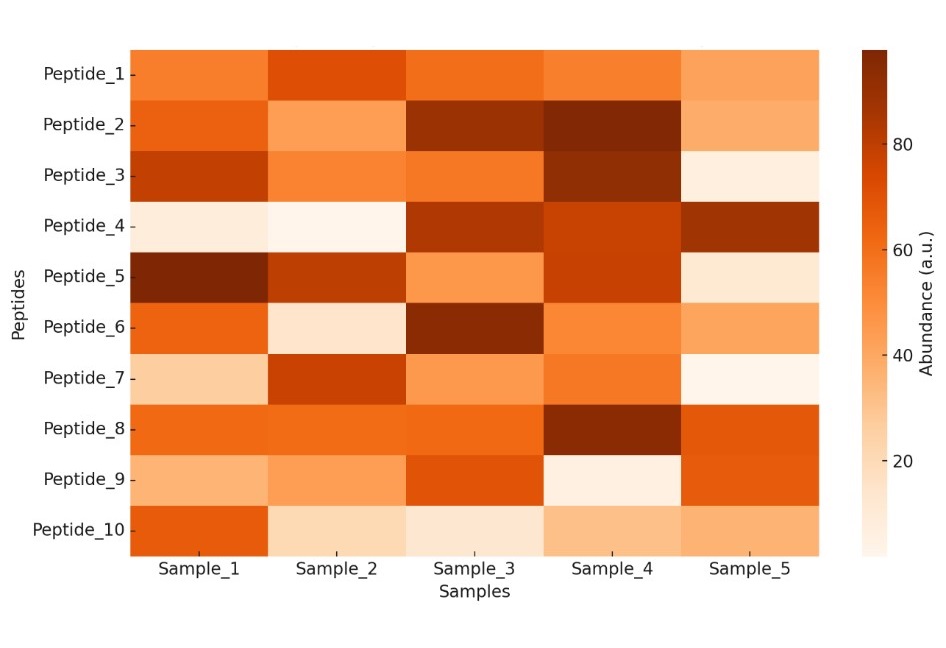
Heatmap of Peptide Abundance Across Samples
Volcano Plot of Differential Peptide Abundance

Deliverables | What You Will Receive
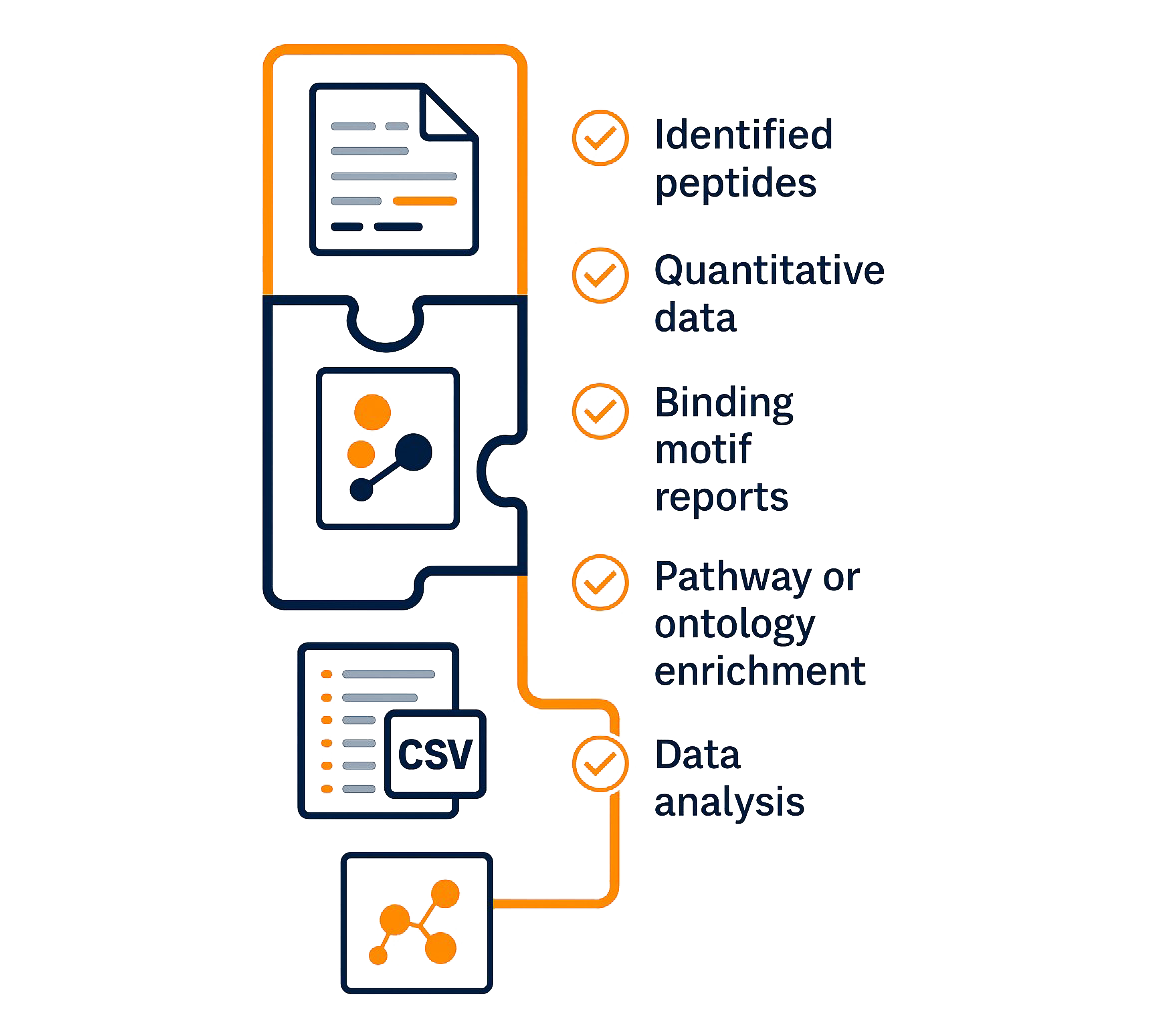
- Identified Peptides (CSV Format)
List of all HLA-bound peptides with sequences, lengths, charge states, and spectral quality scores. - Quantitative Data (Normalized)
Relative peptide abundance (intensity or spectral count) across sample groups, ready for statistical analysis or visualization. - HLA Binding Motif Reports
Sequence logos and binding motif tables derived from deconvoluted peptide sets, categorized by HLA alleles. - PTM and Variant Annotations (if applicable)
Detected phosphorylation, glycosylation, or citrullination sites, and candidate neoantigen peptides with mutation context. - Pathway & Ontology Enrichment (Optional)
Functional insights into peptide source proteins, including GO terms, KEGG pathways, and immune-related processes. - Graphical Summary PDF
A visual executive summary with peptide length distribution, volcano plots, and key motif logos.
FAQ for Immunopeptidome Analysis
Large-Scale Immunopeptidome Analysis Reveals Recurrent Posttranslational Splicing of Cancer- and Immune-Associated Genes
Journal: Molecular & Cellular Proteomics
Published: Volume 22, Issue 4, 2023
Summary
Using advanced mass spectrometry and a novel spliced peptide prediction pipeline, researchers analyzed immunopeptidomic data from cancer patients and cell lines. They identified over 150 recurrent posttranslational spliced peptides presented by HLA class-I and class-II molecules, several of which derived from cancer-related genes like DAPK1 and HLA-E. Importantly, two of these peptides were validated using synthetic standards and shown to
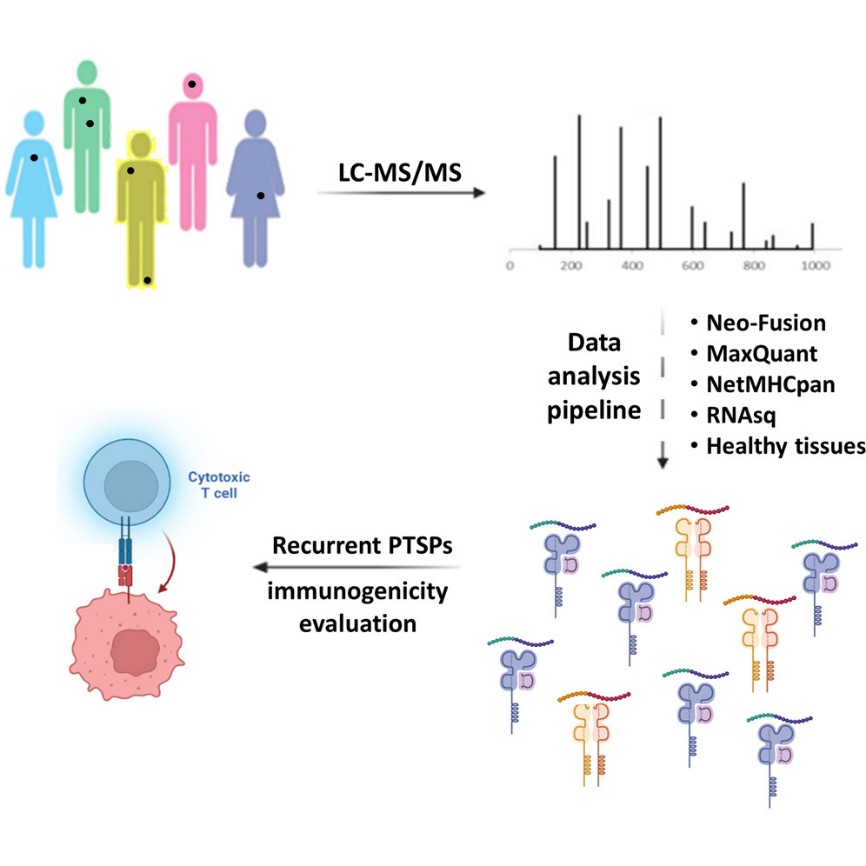
Methods
This study presents a novel computational pipeline combining Neo-Fusion and MaxQuant to stringently identify and validate recurrent PTSPs across multiple cancer types using large-scale MS-based immunopeptidomics data.
Key Technical Features:
- Sample types: Patient-derived melanoma lines, breast cancer (MCF7), HNSCC (SCC47), and mouse melanoma clones
- Peptide identification: Neo-Fusion detects spliced sequences; MaxQuant performs spectrum matching
- Stringent filtration: Peptides were filtered for binding affinity (NetMHCpan), MS/MS coverage, retention time, transcriptomic origin (RNA-seq), and presence in normal or microbial proteomes
- Validation: Light and heavy synthetic peptides were used to confirm MS identifications
- Immunogenicity tests: Conducted using TILs and PBMC-derived T cells
- Recurrent peptides: Defined by presence across multiple independent patients or metastases
Creative Proteomics can offer advanced immunopeptidomics services that mirror and support this research pipeline, including:
- HLA peptidomics profiling using high-resolution LC-MS/MS
- Neoantigen discovery and PTSP identification
- HLA class-I and class-II ligandome mapping
- Quantitative and comparative immunopeptidome analysis across samples or treatment conditions
- Synthetic peptide validation and custom bioinformatics pipelines for spliced peptide filtering
These services empower cancer immunotherapy research, vaccine target screening, and immune evasion mechanism studies.
Results
PTSP Frequency Is Low but Significant
- PTSPs constitute <3.1% of HLA class-I and <0.5% of HLA class-II immunopeptidomes.
- Despite the low proportion, PTSPs outnumber neoantigens derived from somatic mutations, highlighting their potential clinical relevance.
Recurrent PTSPs Identified Across Patients
- 143 HLA class-I and 10 HLA class-II recurrent PTSPs were discovered.
- These peptides are associated with cancer-related genes such as MITF, DAPK1, IL17RE, and HLA-E.
Immunogenicity Validated
- Two PTSPs (from DAPK1 and HLA-E) elicited significant CD8+ T cell responses in autologous TILs and PBMC-derived T cells.
- Flow cytometry showed upregulation of activation markers (e.g., 4-1BB, IFN-γ, TNF-α).
- Dextramer staining confirmed peptide-specific CD8+ T cell populations in healthy donors.
Spliced Peptides Recur in Single-Cell Clones
- Spliced peptides were consistently detected across 5–15 single-cell clones derived from the same mouse tumor, demonstrating biological reproducibility.
Pipeline Validation and Stringency
- Most spurious hits were removed by Delta score and Leu/Ile ambiguity filters.
- Leu/Ile replacement filter alone provided a 30–40× discrimination between WT and spliced peptide false-positives.
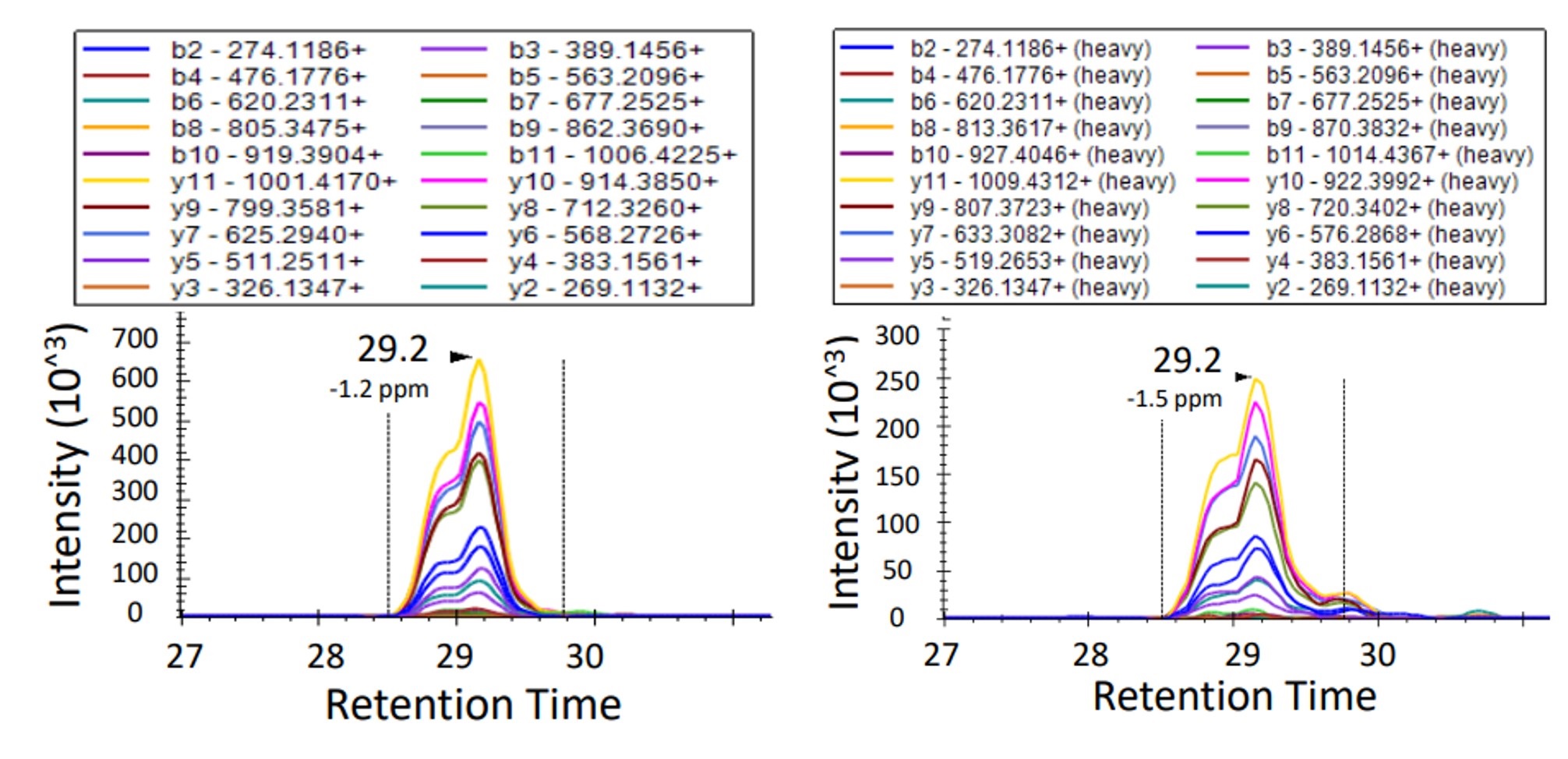 Overlay of endogenous and isotopically labeled HLA-E–derived peptide (WSDSSGGKGGSY) chromatograms in targeted immunopeptidomics of 83T melanoma cells, visualized via Skyline.
Overlay of endogenous and isotopically labeled HLA-E–derived peptide (WSDSSGGKGGSY) chromatograms in targeted immunopeptidomics of 83T melanoma cells, visualized via Skyline.
Reference
- Levy, Ronen, et al. "Large-scale immunopeptidome analysis reveals recurrent posttranslational splicing of cancer-and immune-associated genes." Molecular & cellular proteomics 22.4 (2023). https://doi.org/10.1016/j.mcpro.2023.100519
Combined Omics Approaches Reveal Resistance Mechanisms to Beet Curly Top Virus in Sugar Beet Genotypes
Journal: International Journal of Molecular Sciences
Published: 2023
https://doi.org/10.3390/ijms241915013BRCA1 Antibodies Matter: Benchmarking Reagent Specificity in Cancer Research
Journal: International Journal of Biological Sciences
Published: 2021
https://doi.org/10.7150/ijbs.63115In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1 and Effect of Phorbol Ester on Activation by TcPKC of TcPolβ in Trypanosoma cruzi Epimastigotes
Trypanosoma cruzi DNA Polymerase β Is Phosphorylated In Vivo and In Vitro by Protein Kinase C (PKC) and Casein Kinase 2 (CK2)
The Interplay Between GSK3β and Tau Ser262 Phosphorylation During the Progression of Tau Pathology
Journal: International Journal of Molecular Sciences
Published: 2022
https://doi.org/10.3390/ijms231911610Inactivation of Myeloperoxidase by Benzoic Acid Hydrazide
Journal: Archives of Biochemistry and Biophysics
Published: 2015
https://doi.org/10.1016/j.abb.2015.01.028


 Orbitrap Astral™
Orbitrap Astral™ Orbitrap Exploris™ 480
Orbitrap Exploris™ 480