Vitis vinifera is one of the most economically significant and extensively cultivated fruit crops in the world. It is primarily cultivated for the production of grapes, which are used to make wine, raisins, and various grape-derived products. Understanding the metabolic processes within Vitis vinifera is crucial for enhancing grape and wine quality, improving disease resistance, and optimizing viticultural practices.
Vitis vinifera Metabolomics Analysis Project at Creative Proteomics
Metabolite Profiling: Leveraging cutting-edge mass spectrometry, we offer comprehensive identification and quantification of diverse metabolites within Vitis vinifera samples. From sugars and organic acids to polyphenols and aroma compounds, our metabolite profiling reveals the chemical composition of grapevines.
Metabolic Pathway Analysis: We integrate metabolomics data with pathway databases to illuminate the active metabolic pathways in Vitis vinifera. Gain a deeper understanding of the intricate network of grapevine metabolism and the biological mechanisms driving growth and development.
Biomarker Discovery: Identify potential biomarkers that serve as indicators of grapevine health, stress responses, or specific growth stages. Our metabolomics analysis identifies unique metabolite patterns valuable for grapevine breeding, disease detection, and targeted viticultural strategies.
Comparative Metabolomics: Compare metabolite profiles across different Vitis vinifera cultivars, growth conditions, or developmental stages. Our comparative analysis reveals subtle variations in metabolite levels, helping you comprehend the influence of diverse factors on grapevine metabolism.
Vitis vinifera Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS combines liquid chromatography separation with mass spectrometry detection to identify and quantify metabolites. For example, an Agilent 1290 Infinity LC coupled with an Agilent 6550 iFunnel Q-TOF MS or a Thermo Scientific Q Exactive Series LC-MS/MS system can be utilized.
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is suitable for volatile and semi-volatile compound analysis. Instruments like an Agilent 7890B GC coupled with an Agilent 5977A MSD or a Shimadzu GCMS-QP2020 can be used for metabolite analysis.
Tandem Mass Spectrometry (MS/MS or MS2): MS/MS involves fragmenting ions to gather structural information. For instance, a Thermo Scientific Orbitrap Fusion Tribrid MS or a Waters SYNAPT G2-Si HDMS can be employed for metabolite identification.
High-Resolution Mass Spectrometry: High-resolution mass spectrometry aids in accurate mass measurement. Instruments like an Agilent 6545 Q-TOF LC/MS or a Bruker maXis II ETD provide precise mass analysis for identifying metabolites.
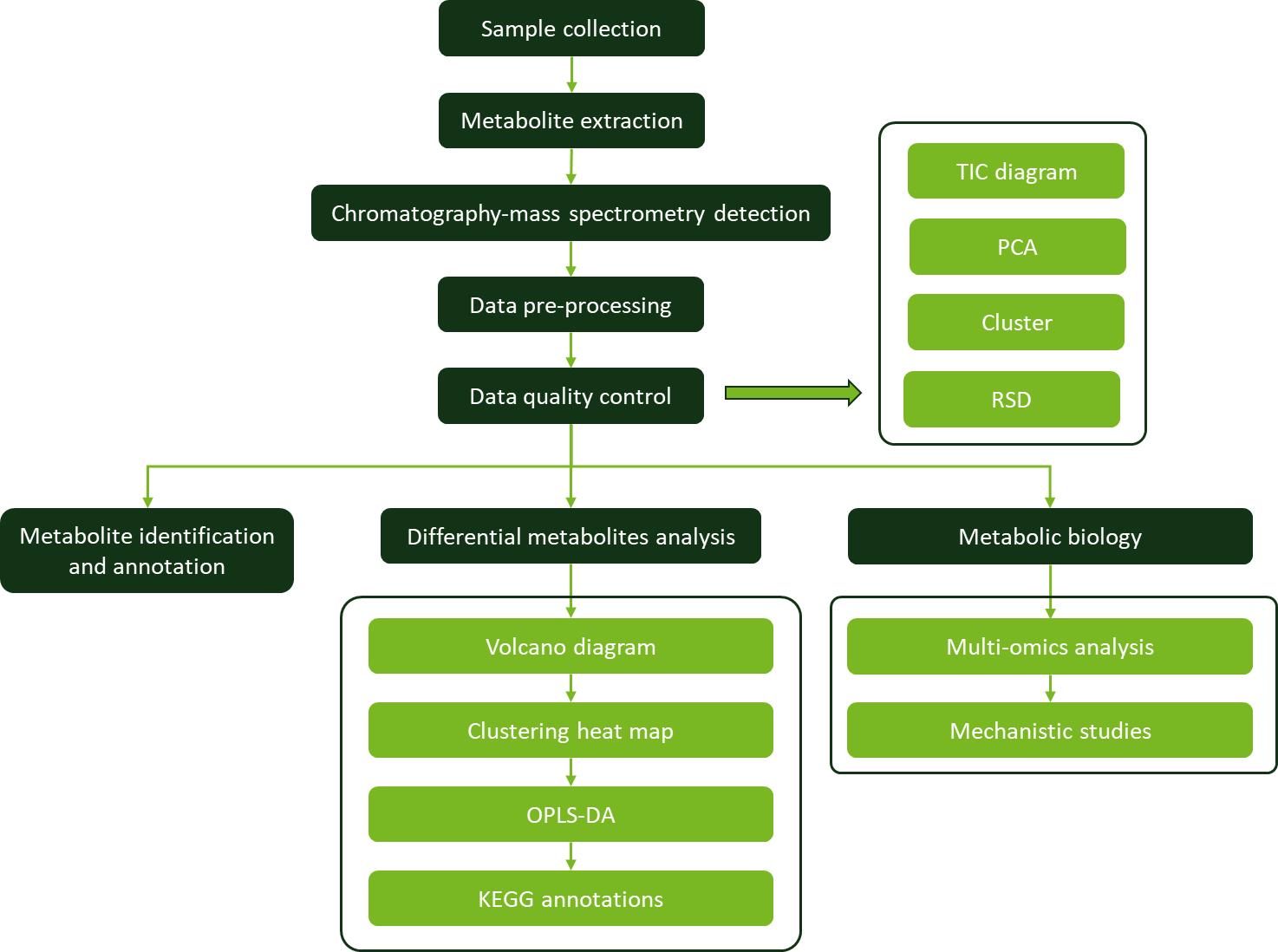 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Applications of Metabolomics in Vitis vinifera Research
1. Analysis of Grape and Wine Quality and Characteristics:
Metabolomics analysis delves into the distinct metabolic differences among various grape varieties and wine types, uncovering pathways responsible for different flavors, aromas, and qualities.
Identification of Specific Flavor and Nutritional Components: Profiling metabolites in grapes and wines helps pinpoint the metabolites associated with specific flavors, aromas, and nutritional attributes.
2. Understanding Environmental Responses:
Metabolomics provides insights into how Vitis vinifera responds to environmental stressors such as temperature changes, water availability, and pathogens.
Identification of Stress-Responsive Metabolites: Metabolomics helps identify metabolites that are altered in response to stress, aiding in the understanding of adaptive mechanisms.
3. Characterizing Growth and Development:
Metabolomics tracks changes in metabolite profiles throughout different growth stages of grapevine, shedding light on the underlying biochemical processes.
Uncovering Developmental Shifts: Metabolomics reveals metabolic shifts during key growth stages, contributing to an understanding of grapevine development.
4. Exploration of Secondary Metabolites:
Secondary metabolites such as phenolic compounds, flavonoids, and terpenes contribute to grape health and wine quality.
Metabolomics uncovers the diversity of secondary metabolites and their roles in defense mechanisms and flavor development.
5. Investigation of Pathway Regulation:
Metabolomics analysis provides insights into metabolic pathways and their regulation in response to changing conditions.
Elucidation of Biosynthetic Pathways: Metabolomics assists in unraveling the biosynthetic routes of compounds like anthocyanins and flavonoids.
6. Identification of Biomarkers:
Metabolomics aids in identifying specific metabolites that act as biomarkers for grapevine health, stress response, or disease susceptibility.
Biomarkers for Disease Detection: Metabolomics helps discover potential markers indicative of disease presence or susceptibility.
7. Optimization of Viticultural Practices:
Metabolomics guides decisions in vineyard management, irrigation strategies, and nutrient supplementation to enhance grape quality.
Evidence-Based Viticulture: Metabolomics data informs strategies for sustainable grape production and quality improvement.
Sample Requirements for Vitis vinifera Metabolomics
| Sample Requirements |
Details |
| Sample Type |
Grapes, grape extracts, juices, wines, leaves, etc. |
| Sample Quantity |
Typically a few grams to milligrams, depending on technique. |
| Sample Homogenization |
Required for plant tissues; grinding, blending, or crushing. |
| Extraction Solvents |
Chosen based on metabolites; common: methanol, acetonitrile, water. |
| Extraction Method |
Solid-liquid extraction, liquid-liquid extraction, QuEChERS, etc. |
| Derivatization (if needed) |
Some metabolites might require derivatization for better detection or stability. |
| Sample Storage |
Cryopreservation with liquid nitrogen or storage at -80°C. |
| Quality Control Samples |
Include internal standards, QC samples for accuracy monitoring. |
Case 1. Genetic Basis and Metabolic Profiling of Grape Stem Polyphenols Revealed Through Targeted Metabolomics
Background:
Grape polyphenols are bioactive compounds with defense and health benefits. This study aimed to explore the metabolic profile of grape stems, which contain these valuable polyphenols. By focusing on genetic determinants and minimizing environmental influences, the study aimed to uncover the genetic basis of polyphenol metabolism.
Samples:
The study conducted a field experiment in the Loire Valley, France, involving eight grape varieties: Sauvignon, Chenin, Chardonnay, Pinot Noir, Grolleau, Gamay, Malbec, and Cabernet Franc. The experiment utilized adjacent vineyard plots with the same soil type to minimize environmental variability.
Methods:
The study employed a UPLC-MS-based targeted metabolomic approach to analyze the polyphenol composition of grape stems from different genotypes. The detailed procedure consisted of the following steps:
Sample Collection and Preparation:
Vineyards in the Loire Valley were selected, with adjacent plots having the same calcareous clay soil. Grape stems from eight different grape varieties were collected during grape vine dormancy to minimize the impact of environmental factors. To mitigate intra-plot variation, grape stems were randomly pruned from different parcels within each plot. This sampling strategy aimed to capture the genetic variation in polyphenol accumulation.
Metabolite Extraction:
The collected grape stems were subjected to metabolite extraction to obtain the polyphenolic compounds. The extraction process involved homogenization of the plant material followed by appropriate solvent extraction to solubilize the metabolites.
UPLC-MS Analysis:
The extracted metabolites were then subjected to UPLC-MS analysis, which stands for Ultra Performance Liquid Chromatography-Mass Spectrometry. This technique allows for the separation, identification, and quantification of individual compounds within a complex mixture based on their mass and retention time.
Targeted Metabolomic Analysis:
The UPLC-MS analysis was targeted specifically at polyphenols, with a focus on stilbenoids and related compounds. Calibration curves using pure standards of known polyphenols were used to quantify the major stilbenoids present in the samples. The UPLC-MS analysis provided information on the mass-to-charge ratio (m/z) of the molecular ions in the samples.
Molecular Assignment and Annotation:
The UPLC-MS data were analyzed to identify and annotate the polyphenolic compounds present in the samples. This involved comparing the observed molecular ions with those of known standards and utilizing data from previous studies. Compounds were provisionally assigned based on mass data, UV spectra, and elution order.
Multivariate Statistical Analysis:
To analyze the data comprehensively, multivariate statistical techniques were applied. Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) were used to visualize similarities and differences between samples based on their polyphenolic profiles. Hierarchical clustering analysis (HCA) was also conducted to identify distinct groups within the dataset.
Results
The study revealed significant variations in major stilbenoids among the selected grape genotypes. It identified E-resveratrol, E-ε-viniferin, ampelopsin A, and hopeaphenol as the major stilbenoids. A detailed UPLC-MS analysis identified 42 compounds in total, including stilbenoids and other polyphenols. Multivariate statistical analyses allowed discrimination of samples based on both berry color and genotype. Hierarchical clustering and correlation-based network analysis highlighted the relationship between genetic distance and metabolomic differences.
By focusing on genetic determinants of polyphenol metabolism and utilizing a targeted metabolomic approach, this study provided insights into the complexity of grape stem metabolomes and their relationship to genetic background and biochemical pathways.
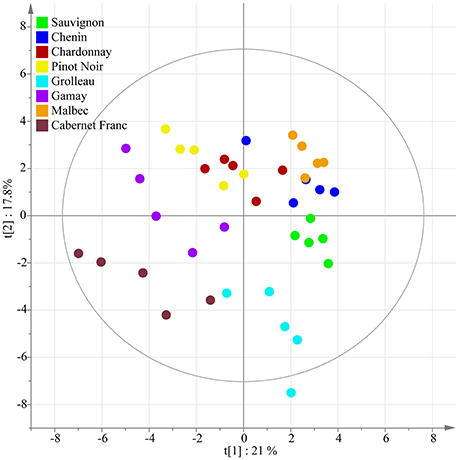 Unsupervised classification using PCA.
Unsupervised classification using PCA.
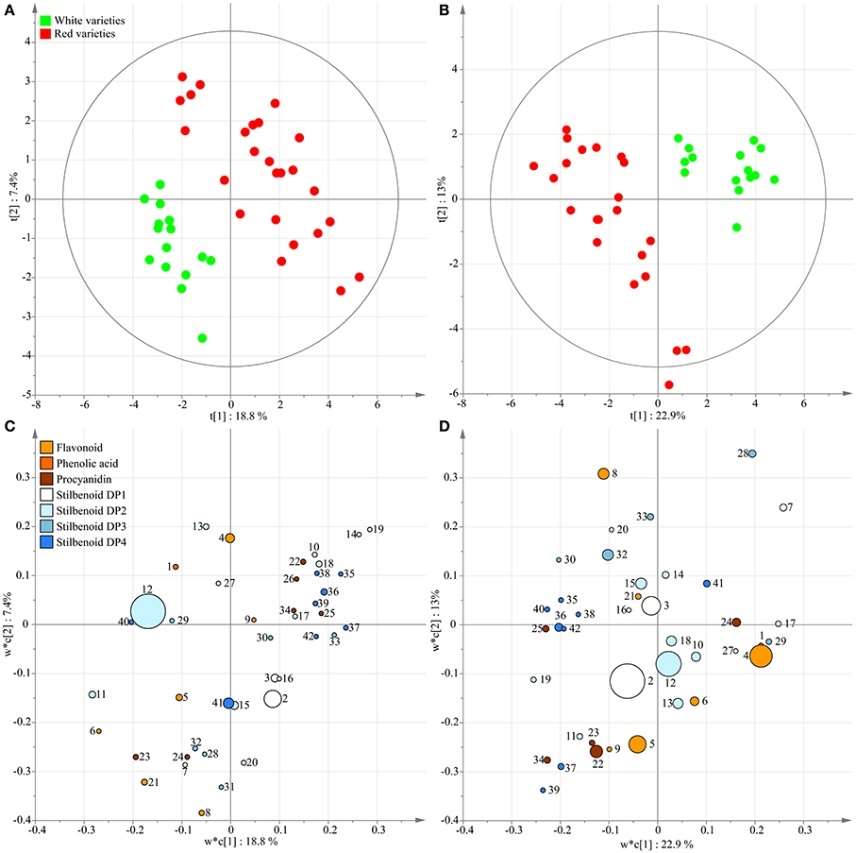 Supervised classification using PLS-DA.
Supervised classification using PLS-DA.
Reference
- Billet, Kévin, et al. "Field-based metabolomics of Vitis vinifera L. stems provides new insights for genotype discrimination and polyphenol metabolism structuring." Frontiers in plant science 9 (2018): 798.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Unsupervised classification using PCA.
Unsupervised classification using PCA. Supervised classification using PLS-DA.
Supervised classification using PLS-DA.