Arabidopsis thaliana as a Model Organism
Arabidopsis thaliana, commonly referred to as Arabidopsis, is a small flowering plant belonging to the mustard family (Brassicaceae). Arabidopsis possesses several characteristics that make it an ideal model organism for metabolomics research. Firstly, its relatively simple and compact genome of about 125 million base pairs makes it a tractable subject for genetic manipulation and analysis. This simplicity accelerates the identification of metabolic pathways and enables researchers to pinpoint the roles of specific genes in metabolic processes.
Secondly, Arabidopsis has a rapid life cycle, with a complete growth and reproduction cycle taking as little as six weeks. This rapid turnover allows researchers to conduct experiments and observe changes in metabolite profiles over multiple generations in a relatively short time span.
Moreover, Arabidopsis shares genetic homology with many crop species, making it a valuable reference point for understanding the metabolic processes of economically important plants. Insights gained from Arabidopsis metabolomics studies can be applied to enhance crop yield, quality, and resistance to various stressors, thereby contributing to global agricultural sustainability.
The metabolic pathways in plants are intricate networks of chemical reactions that govern the synthesis and breakdown of molecules essential for growth, development, and defense. Arabidopsis serves as a valuable tool for unraveling these pathways due to its amenability to various experimental techniques and its comprehensive genetic resources.
The information obtained from Arabidopsis metabolomics studies aids in identifying key metabolites, enzymes, and regulatory mechanisms involved in essential processes such as photosynthesis, hormone signaling, and secondary metabolite production.
Arabidopsis Metabolomics Analysis at Creative Proteomics
At Creative Proteomics, our Arabidopsis metabolomics services are tailored to leverage the unique attributes of this model organism. Our advanced analytical techniques, such as gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS), allow for the comprehensive profiling of metabolites in Arabidopsis samples. We offer both targeted and untargeted metabolomics approaches, enabling us to explore known metabolic pathways and discover novel metabolites, respectively.
Our skilled team of experts collaborates with researchers to design experiments, analyze data, and interpret results, ensuring that valuable insights are extracted from each study. By offering precise and insightful metabolomics analysis, we contribute to the broader scientific community's understanding of Arabidopsis metabolism and its implications for plant biology.
Targeted Metabolite Profiling:
Our targeted metabolomics approach focuses on the quantitative analysis of specific metabolites within the Arabidopsis metabolome. This solution is ideal for researchers aiming to explore particular metabolic pathways or assess the concentration of key metabolites in response to different stimuli or genetic modifications.
Untargeted Metabolomics Analysis:
Our untargeted metabolomics solution involves a comprehensive profiling of the entire Arabidopsis metabolome without any prior bias. This approach is particularly beneficial for discovering novel metabolites, identifying unexpected pathway interactions, and obtaining a holistic view of metabolic changes.
Metabolic Pathway Analysis:
Our metabolic pathway analysis solution involves mapping metabolomics data onto known metabolic pathways to enable clear visualization of metabolic interactions, helping you to identify key regulators, metabolic bottlenecks, and potential targets for manipulation.
Comparative Metabolomics Studies:
Comparative metabolomics allows comparison of metabolite profiles between different Arabidopsis samples, such as wild-type and mutant plants, treated and untreated samples, or samples exposed to different environmental conditions, to identify metabolites responsible for specific phenotypic features and to reveal the molecular mechanisms underlying these differences.
Customized Metabolomics Services:
At Creative Proteomics, we understand that research questions are diverse and unique. Therefore, we offer customized Arabidopsis metabolomics services tailored to your specific research needs. Our team of experts collaborates closely with you to design experiments, select appropriate analytical techniques, analyze data, and provide insightful interpretations, ensuring that your research goals are met effectively.
Arabidopsis Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS)
Our LC-MS analyses are conducted using instruments such as the Agilent 1290 Infinity II LC System coupled with the Agilent 6550 iFunnel Q-TOF LC/MS. This powerful combination allows us to achieve unparalleled resolution and sensitivity. The LC system efficiently separates metabolites, while the Q-TOF mass spectrometer accurately determines the molecular masses and fragmentation patterns of detected compounds. This setup enables the comprehensive profiling of a wide range of metabolites, from polar to nonpolar compounds, contributing to a holistic understanding of Arabidopsis metabolism.
Gas Chromatography-Mass Spectrometry (GC-MS)
For GC-MS analyses, we employ the Thermo Scientific™ TRACE™ 1310 Gas Chromatograph coupled with the Thermo Scientific™ ISQ™ Series Single Quadrupole Mass Spectrometer. This robust pairing excels in the analysis of volatile and thermally stable metabolites. The GC system separates compounds based on their volatility, and the mass spectrometer identifies and quantifies the metabolites with high precision. Through this approach, we capture key plant-specific compounds that play pivotal roles in growth, defense, and adaptation.
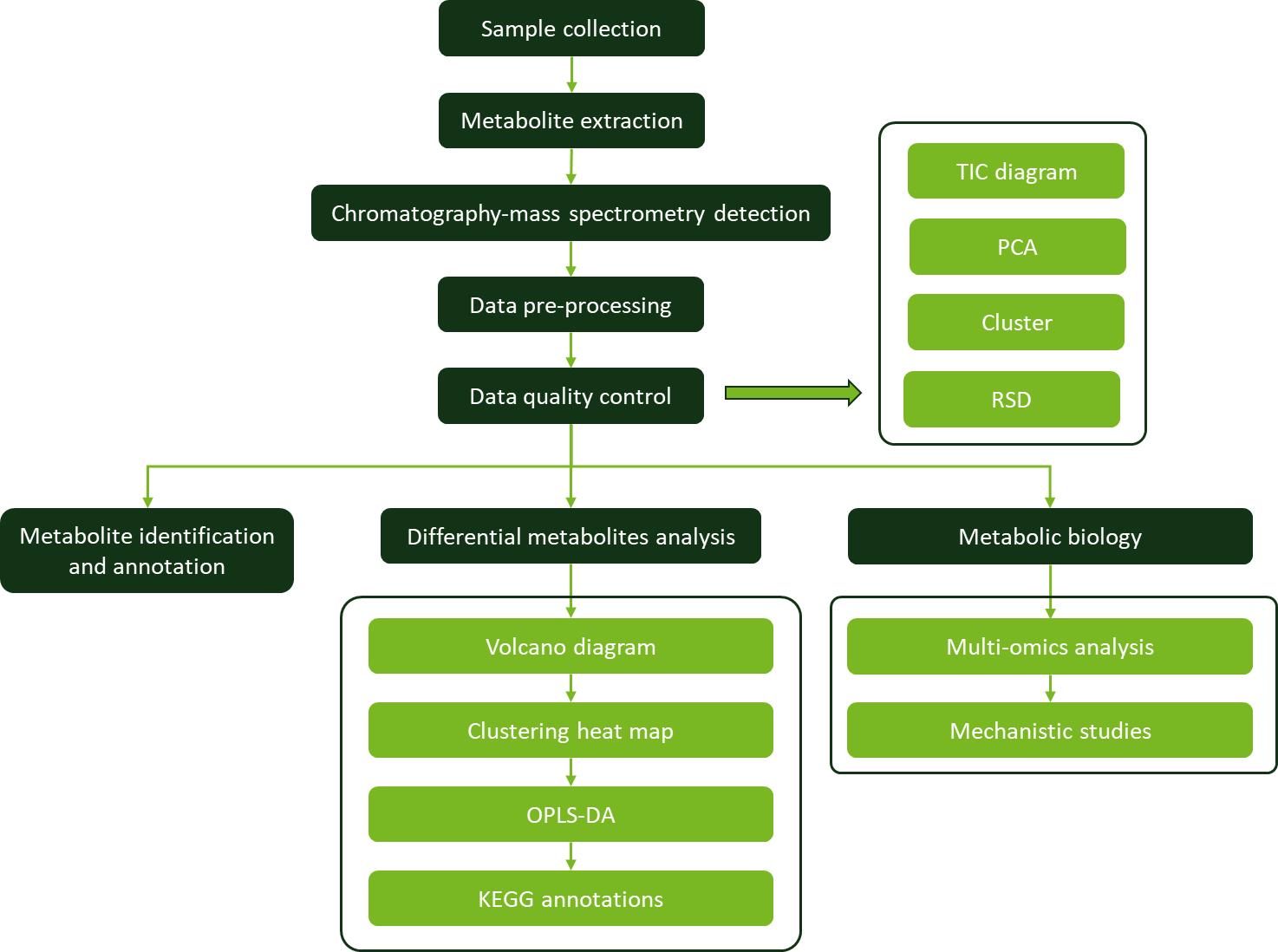 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Data Analysis and Interpretation
| Step |
Description |
| Preprocessing |
- Data cleaning to remove noise
- Retention time alignment
- Data quality assessment |
| Feature Extraction |
- Identification of detected features
- Annotation of features against databases |
| Data Normalization |
- Correction for technical variations
- Sample concentration adjustment |
| Statistical Analysis |
- Univariate tests for individual metabolites
- Identification of significantly changed metabolites |
| Multivariate Analysis |
- PCA and PLS-DA to reveal patterns and relationships in the data |
| Pathway Enrichment |
- Mapping metabolites onto known pathways
- Identification of overrepresented pathways |
| Biological Interpretation |
- Integration of statistical results and pathway information
- Hypothesis generation |
| Visualization |
- Heatmaps, volcano plots, pathway diagrams
- Data visualization for communication |
Sample Requirements for Arabidopsis Metabolomics
| Sample Type |
Typical Sample Quantity |
| Leaf Tissue |
100 - 200 mg |
| Root Tissue |
100 - 200 mg |
| Stem Tissue |
100 - 200 mg |
| Flower Tissue |
50 - 100 mg |
| Seed Tissue |
20 - 50 mg |
Case 1. Exploring Arabidopsis Diversity and Defense Mechanisms Through Multi-Platform Metabolomics
Background:
Metabolomics is a powerful tool for studying the intricate relationships between a plant's genetics, its metabolism, and its response to the environment. Arabidopsis thaliana, a well-studied model plant, offers an excellent opportunity to explore these connections due to its genetic diversity and susceptibility to various biotic agents.
Samples:
Nine accessions of Arabidopsis thaliana were selected for this study. These accessions represented a range of genetic diversity and were chosen to investigate the metabolic variations within the species. The goal was to uncover how genetic differences lead to distinct metabolic profiles, particularly in the context of plant defense responses against biotic agents.
Methods:
The researchers employed a multi-platform metabolomics approach to comprehensively profile the metabolome of the Arabidopsis accessions. This approach involved the integration of liquid chromatography coupled with time-of-flight mass spectrometry (LC-TOF-MS) in both positive and negative ionization modes, along with gas chromatography coupled with time-of-flight mass spectrometry (GC-TOF-MS). The utilization of multiple methods enabled a broader coverage of metabolites and their chemical characteristics.
The LC-TOF-MS analysis provided detailed information about a wide range of metabolites, including glucosinolates and phenolic compounds. The negative mode analysis confirmed the identity of various glucosinolates, while a novel phenolic compound was identified in the negative mode as well. In the positive mode, alkaloids and flavonoid derivatives were detected, some of which were not previously reported in Arabidopsis.
Results
The study revealed significant metabolic diversity among the nine Arabidopsis accessions. This diversity was influenced by both genetic factors and the interaction with biotic agents. Interestingly, the metabolic distance did not always align with genetic distance, indicating the complex interplay between genetics and the environment in shaping the metabolome.
The researchers also found specific metabolites that correlated with the plants' responses to various biotic agents, suggesting that metabolic diversity plays a role in defense mechanisms. The integration of metabolomics data with genetic information led to insights into how certain compounds are linked to specific interactions with biotic agents.
In conclusion, this study showcased the utility of integrative metabolomics techniques to unravel the intricate connections between genetics, metabolism, and environmental interactions in Arabidopsis thaliana. The results shed light on the complex relationship between genetic diversity and metabolic diversity and provided new insights into the plant's defense strategies against biotic stressors.
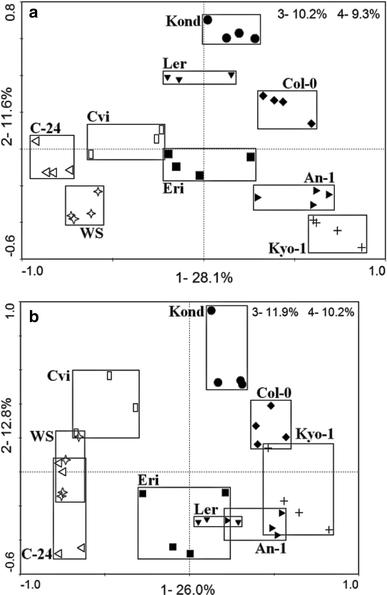 PCA scores plots of the root metabolite profile of nine accessions grown in hydroponics, analyzed by LC–MS in positive mode (a) and negative mode (b).
PCA scores plots of the root metabolite profile of nine accessions grown in hydroponics, analyzed by LC–MS in positive mode (a) and negative mode (b).
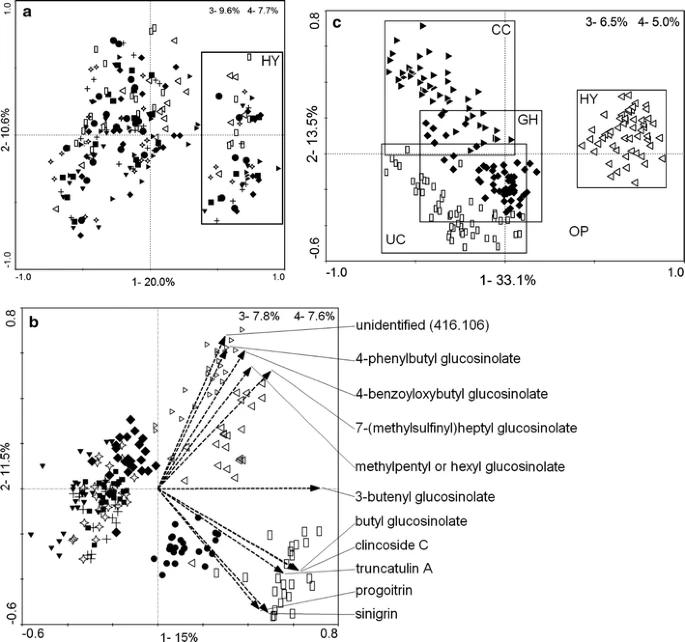 PCA plots of the nine accessions grown in four environments, analyzed by LC–MS of shoot in negative-mode.
PCA plots of the nine accessions grown in four environments, analyzed by LC–MS of shoot in negative-mode.
Reference
- Houshyani, Benyamin, et al. "Characterization of the natural variation in Arabidopsis thaliana metabolome by the analysis of metabolic distance." Metabolomics 8 (2012): 131-145.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service PCA scores plots of the root metabolite profile of nine accessions grown in hydroponics, analyzed by LC–MS in positive mode (a) and negative mode (b).
PCA scores plots of the root metabolite profile of nine accessions grown in hydroponics, analyzed by LC–MS in positive mode (a) and negative mode (b). PCA plots of the nine accessions grown in four environments, analyzed by LC–MS of shoot in negative-mode.
PCA plots of the nine accessions grown in four environments, analyzed by LC–MS of shoot in negative-mode.