What are Plants Targeted Metabolomics?
Plant targeted metabolomics involves the systematic identification and quantification of a specific subset of metabolites within plant tissues. These metabolites play vital roles in plant growth, development, and responses to external stimuli, making their precise analysis crucial for understanding plant physiology and biochemistry. At Creative Proteomics, we offer plant targeted metabolomics analysis to unravel the complex metabolic profiles of plants, enabling researchers to decipher the underlying mechanisms driving plant biology.
Specific Projects in Plants Targeted Metabolomics
Plant targeted metabolomics can provide valuable insights into various metabolic pathways, plant responses to stress, and the identification of biologically active compounds. Creative Proteomics offers the following specific programs in plant targeted metabolomics:
- Understanding plant-pathogen interactions:
Using targeted metabolomics to study how plants respond to pathogen attack. Gain insight into the molecular mechanisms of plant resistance by quantifying specific metabolites involved in defense pathways, such as phytoestrogens and secondary metabolites.
- Medicinal Plant Research:
Targeted metabolomics can be used to identify and quantify bioactive compounds in medicinal plants, which can help identify potential natural drug products and help optimize cultivation conditions to improve yields of specific metabolites.
- Crop improvement and breeding:
Targeted metabolomics is utilized to identify metabolites associated with desirable traits such as nutrient content, disease resistance, and abiotic stress tolerance to guide breeding efforts to develop improved crop varieties.
- Abiotic stress response studies:
Targeted metabolomics can reveal how plants respond to environmental stresses such as drought, salinity and temperature extremes. By quantifying stress-related metabolites, metabolic adaptations that allow plants to survive and thrive under challenging conditions are revealed.
- Interactions between plants and endophytes:
Targeted metabolomics can be used to study the interactions between plants and endophytic microbes. By analyzing specific metabolites produced by plants and their endosymbiotic microbes, it is possible to discover symbiotic relationships and the potential benefits these microbes provide.
- Metabolic engineering for biofuel production:
Targeted metabolomics is applied to engineer metabolic pathways in plants to increase biofuel production. Optimizing plant biochemistry by manipulating specific metabolites involved in energy storage and conversion leads to more efficient biofuel production.
- Secondary metabolite regulation:
Targeted metabolomics helps to elucidate the mechanisms regulating the biosynthesis and accumulation of secondary metabolites such as flavonoids, alkaloids, and terpenoids, and provides a basis for strategies to increase the production of valuable secondary metabolites.
- Ectopic and plant competition:
Studying allelopathic interactions between plants, where certain metabolites released by one plant inhibit the growth of neighboring plants. Quantifying allelopathic compounds can help understand plant competition and ecosystem dynamics.
Plants Targeted Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS is one of the most widely employed techniques for targeted metabolomics. It separates metabolites in a sample using liquid chromatography based on their chemical properties and then quantifies and detects them using mass spectrometry.
Instrument Model: Agilent 6460 Triple Quadrupole LC/MS
- Fragmentation Mode: Collision-Induced Dissociation (CID)
- Sensitivity: Sub-femtomole to low picomole range
Instrument Model: Thermo Scientific Q Exactive Series
- Fragmentation Mode: Higher-energy Collisional Dissociation (HCD)
- Sensitivity: Low femtomole range
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is commonly used for volatile and thermally stable metabolites. It vaporizes and separates metabolites in a sample using gas chromatography, followed by mass spectrometric detection.
Instrument Model: Agilent 7890A GC/5975C MSD
- Fragmentation Mode: Electron Impact (EI)
- Sensitivity: Low picogram to nanogram range
Instrument Model: Thermo Scientific TSQ 8000 GC-MS
- Fragmentation Mode: Electron Impact (EI)
- Sensitivity: Low picogram to nanogram range
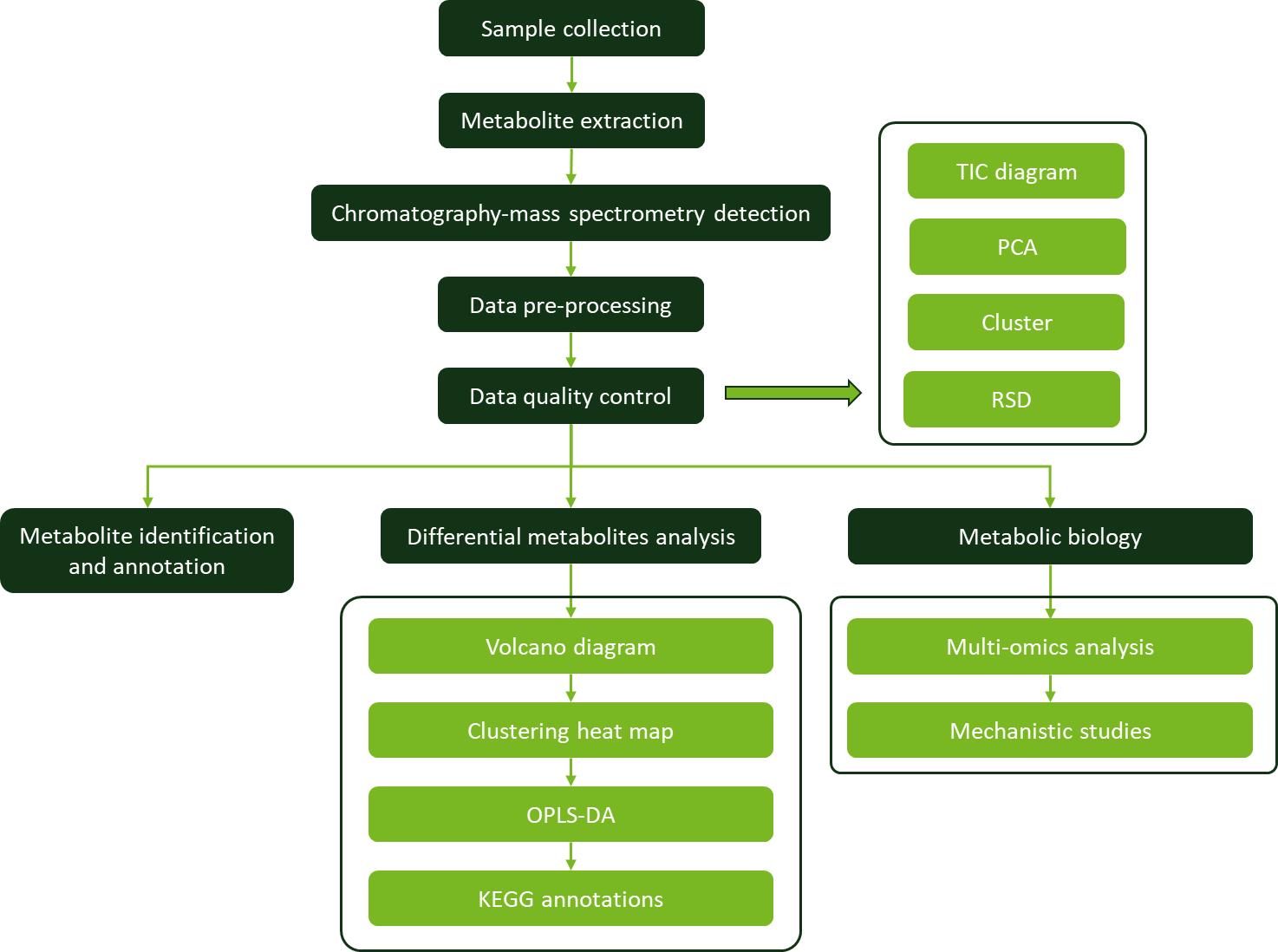 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Our Plant Targeted Metabolism Database (including but not limited to)
| Substance Category |
Compounds |
Quantity |
| Amino acids |
Threonine, Serine, Tyrosine, Valine, Glutamine, Cysteine, etc. |
230+ |
| Vitamins |
Vitamin A, Vitamin B12, Vitamin C, Vitamin D2, Vitamin K1, etc. |
60+ |
| Organic acids |
Fenugreek Acid, Malic Acid, Mangiferic Acid, Ferulic Acid, Citric Acid, Tartaric Acid, etc. |
200+ |
| Carbohydrates |
Lactose, Galactose, Sorbitol, Xylitol, Mannitol, Arabinose, etc. |
50+ |
| Lipids |
Linoleic Acid, Palmitic Acid, Linolenic Acid, Myristic Acid, Arachidonic Acid, etc. |
270+ |
| Nucleotides |
Dimethylguanosine, 2'-Deoxyguanosine, 5-Methylcytosine, Adenine, Adenosine, Cytosine, Guanine, 2-Deoxyadenosine, Pseudouridine, etc. |
150+ |
| Phenols |
Rhododendron, Rhizophorin, Salvinorin, Curcumin, Rhodiol Glycosides, etc. |
80+ |
| Phenolic Acids |
Caffeic Acid, Catechin Gallate, Benzoylpaeoniflorin, Salvinorin A, etc. |
450+ |
| Flavonoids |
Baicalein, Baicalein, Luteolin, Populin, Kaempferol, Proanthocyanidin B1, etc. |
1000+ |
| Alkaloids |
Picrasidine, Rifampicin, Betaine, Ricin, Crotonin, etc. |
600+ |
| Terpenes |
Astragaloside I, Ginsenoside, Betulinol, Carvone, etc. |
350+ |
| Quinones |
Dihydrotanshinone I, Oryzanthin, Rhodopsin, Tanshinone lIIA, Chrysin, etc. |
160+ |
| Steroids |
Phytosterols, Lycorine, Ophiopogonin D, Digitalis Saponin, Allium Veratrum, Riluzin, etc. |
200+ |
| Phenylpropanoids |
Geranylgeranyl Glucoside, Tanshinoside, Chlorogenic Acid, 1-Caffeoylquinic Acid, etc. |
150+ |
| Lignin |
Sinapogenin, Schisandra Chinensis Ester A, Schisandra Chinensis Phenol, Turpentine Glucoside, etc. |
180+ |
| Coumarin |
Coumarin, Coumarin Estrogens, Qiangshuol, Peppercorn Toxin, Scopolamine Glycosides, etc. |
180+ |
| Tannin |
Ephedra Dione, Brown Catechin, Cinnamon Tannin, Cinnamon Tannin, etc. |
140+ |
| Essential Oils |
Limonene, Linalool, Pinene, etc. |
30+ |
| Others |
Bamboo Rhododendron, Xanthophyllin B, Acetobenzene, Patchouli Ketone, etc. |
400+ |
Sample Requirements for Plants Targeted Metabolomics Assay
| Sample Types |
Minimum Sample Size |
| Plant Samples |
Roots, stems and leaves, floral parts, fruits/seeds, rhizomes, buds/tender leaves, tissue sections, pollen, bark, trunk/wood, resin/gum, resin acids, seedlings/young plants, rhizosphere soil, root exudates. |
50 mg - 1 g |
Plants Targeted Metabolomics vs. Untargeted Metabolomics
| Aspects |
Plant Targeted Metabolomics |
Plant Untargeted Metabolomics |
| Purpose |
Precisely quantify specific metabolites of interest |
Explore the complete metabolite landscape without bias |
| Sample Preparation |
Requires prior knowledge of target compounds |
No prior knowledge needed, allows for unbiased analysis |
| Analysis Approach |
Employ specific assays for target metabolites |
Utilize high-resolution techniques for broad spectrum analysis |
| Data Analysis |
Focuses on quantifying pre-defined metabolites |
Involves complex data mining for holistic metabolite profiling |
| Metabolite Identification |
Based on comparison with known standards |
Relies on spectral matching and databases for compound identification |
| Key Advantages |
Accurate quantification, targeted insight |
Discovery of novel compounds and broader metabolic insights |
| Application Examples |
Investigate plant responses to specific stressors |
Uncover diverse metabolic changes under varying conditions |
Case 1. Targeted Metabolomic Profiling Reveals Evolutionary Insights into Signaling Lipids Across Diverse Plant Species
Background:
The study aimed to explore the distribution of specific lipids, including juniperoyl ethanolamide (JEA) and 1/2-juniperoyl glycerol (1/2-JG), across a diverse range of plant species. These lipids, known to play roles in signaling and defense in animals, were investigated within the context of plant evolution and chemotaxonomy. The presence and roles of these lipids in different plant groups were of particular interest.
Samples:
A selection of plant species was chosen based on phylogenetic considerations, covering a wide spectrum of plant types, including algae, lichens, mosses, ferns, gymnosperms, and angiosperms. A total of 71 plant species samples were collected from various sources, encompassing botanical gardens, field collections, and laboratory-grown plants. Leaves were the primary plant material studied, with some cases involving whole plants due to challenges in leaf separation.
Methods:
Synthesis of Juniperoyl Ethanolamide (JEA): JEA was synthesized through a series of chemical reactions involving JuA, oxalyl chloride, DMF. The process was carried out under an argon atmosphere, with temperature control to prevent oxidation. The resulting product was purified using HPLC, and its structure was confirmed using NMR spectroscopy and MS.
Synthesis of 1/2-Juniperoyl Glycerol (1/2-JG): Similar to JEA synthesis, 1/2-JG was obtained from JuA through a multi-step process, including reaction with oxalyl chloride, DMF, cis-1,3-O-benzylideneglycerol, and B-chlorocatecholborane. The mixture of isomers was purified using HPLC, and NMR spectroscopy and MS confirmed their structures.
Plant Selection, Collection, and Preparation: Plant species were carefully selected to represent different phylogenetic groups. Leaves were collected from various locations and extracted with dichloromethane (DCM). The resulting DCM extracts were dried, stored, and later reconstituted in acetonitrile (ACN) for analysis.
Quantification and Identification by LC-MS/MS and GC-MS: Mass spectrometric analysis was conducted using an API 4000 QTrap connected to a Shimadzu UFLC system. Target compounds, including JEA, 1/2-JG, AA, AEA, 1/2-AG, MEA, DHGLA, ScA, ALA, GLA, and JA, were quantified and identified using specific chromatographic conditions, gradients of solvents, and multiple reaction monitoring (MRM) transitions. Additionally, GC-MS analysis was employed for certain compounds.
Multivariate Data Analysis (PCA): Principal Component Analysis (PCA) was performed using SIMCA© software. Autoscaling was applied as a preprocessing step to explore relationships and patterns in the lipid profiles of the diverse plant samples.
Results
The investigation revealed distinctive metabolic profiles associated with different plant groups, such as angiosperms, gymnosperms, monilophytes, mosses, and algae. Notably, the presence of JEA and 1/2-JG, derived from JuA, was identified in specific plant species across gymnosperms and some monilophytes. The distribution of arachidonic acid (AA) and related compounds varied among plant groups, and the absence of AA in angiosperms was noteworthy. The study shed light on potential chemotaxonomic relationships and evolutionary implications of these signaling lipids in the plant kingdom. Further research is suggested to explore the roles of these lipids in plants and their potential interactions with various plant physiological processes.
Overall, the study provided insights into the distribution, synthesis, and potential functions of specific lipid compounds across diverse plant species, contributing to a better understanding of the evolutionary and chemotaxonomic aspects of these molecules in the plant kingdom.
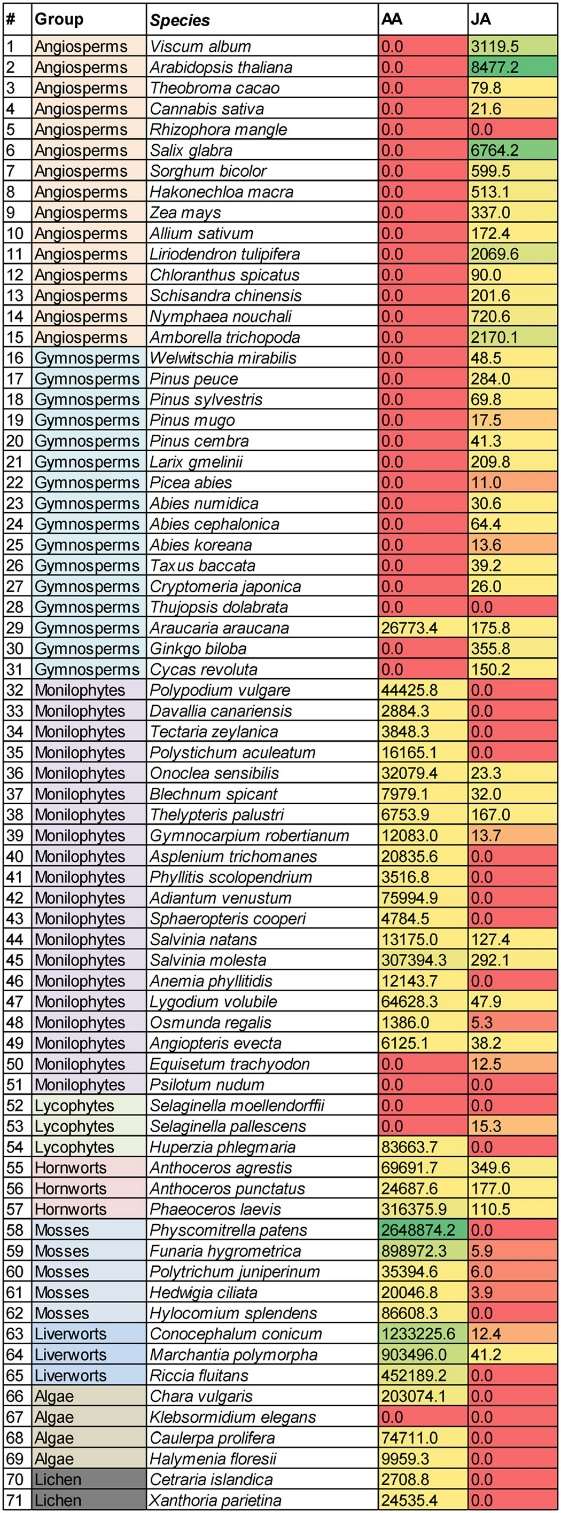 Concentrations (mean values) of AA and JA in pmol/g plant weight found in the 71 plant species analyzed (0.0 = < LOD).
Concentrations (mean values) of AA and JA in pmol/g plant weight found in the 71 plant species analyzed (0.0 = < LOD).
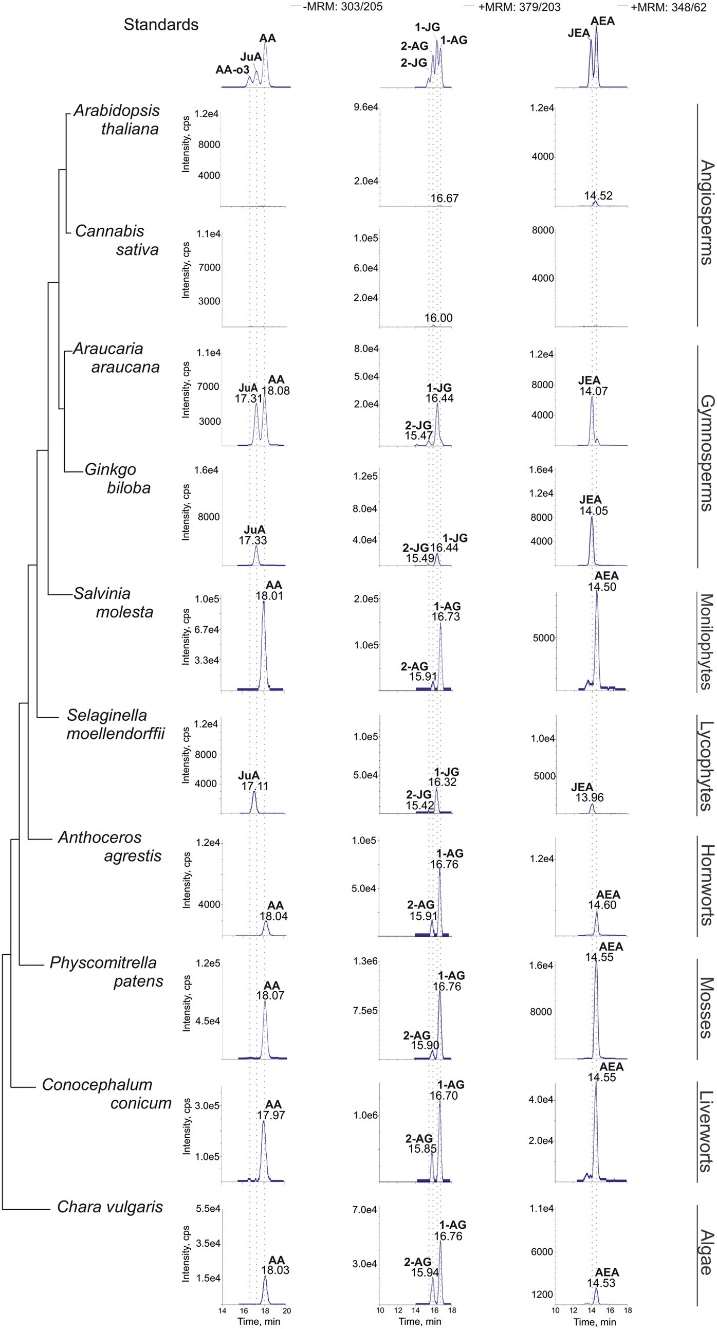 Chromatograms showing the analysis of C20 PUFA metabolites.
Chromatograms showing the analysis of C20 PUFA metabolites.
Reference
- Gachet, María Salomé, et al. "Targeted metabolomics shows plasticity in the evolution of signaling lipids and uncovers old and new endocannabinoids in the plant kingdom." Scientific reports 7.1 (2017): 41177.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Concentrations (mean values) of AA and JA in pmol/g plant weight found in the 71 plant species analyzed (0.0 = < LOD).
Concentrations (mean values) of AA and JA in pmol/g plant weight found in the 71 plant species analyzed (0.0 = < LOD). Chromatograms showing the analysis of C20 PUFA metabolites.
Chromatograms showing the analysis of C20 PUFA metabolites.