What are Plant Natural Product Biosynthetic Pathways?
Plant natural product biosynthetic pathways represent the intricate and highly regulated sequences of biochemical reactions within plant cells that lead to the production of a diverse array of secondary metabolites. These metabolites, also known as natural products, are organic compounds that are not directly involved in the plant's growth, development, or reproduction but often play crucial roles in the plant's adaptation to its environment and interaction with other organisms.
The biosynthetic pathways responsible for generating plant natural products are complex and multifaceted, involving numerous enzymes, intermediates, and cellular compartments. These pathways have evolved over millions of years, allowing plants to synthesize a vast array of compounds with various structures and functions.
Key Components of Plant Natural Product Biosynthetic Pathways:
- Precursor Molecules: The pathways often start with simple precursor molecules derived from primary metabolism, such as amino acids, sugars, and organic acids.
- Enzymatic Reactions: Enzymes catalyze a series of specific reactions, transforming precursor molecules into intermediate compounds. These enzymes are often encoded by specific genes within the plant's genome.
- Compartmentalization: Biosynthetic pathways are often compartmentalized within different cellular organelles, such as the endoplasmic reticulum, chloroplasts, or mitochondria. This spatial organization helps to control the flow of intermediates and ensures the efficiency of the synthesis process.
- Regulation: The expression of genes encoding enzymes involved in biosynthetic pathways is tightly regulated. Environmental factors, developmental cues, and external stimuli can influence the activation or repression of these genes, modulating the production of specific natural products.
- Diversity of Natural Products: Plant natural product biosynthetic pathways give rise to a remarkable diversity of compounds, including alkaloids, flavonoids, terpenoids, and polyphenols. Each class of natural products has distinct chemical structures and biological functions.
The significance of plant natural products extends far beyond the boundaries of botanical research. In medicine, these compounds serve as a treasure trove for the development of pharmaceuticals. In agriculture, they play vital roles in plant defense mechanisms against pathogens and pests. Additionally, industries leverage these compounds for the production of flavors, fragrances, and various other valuable chemicals.
At Creative Proteomics, we recognize the paramount significance of metabolomics research in delving into the intricate world of plant natural product biosynthetic pathways.
Plant Biosynthetic Pathways Metabolomics Solutions
 Dynamic Metabolomics Analysis:
Dynamic Metabolomics Analysis:
Our state-of-the-art metabolomics platform offers a dynamic analysis of plant metabolites, providing a real-time view of biosynthetic pathways. This enables a nuanced understanding of how these pathways respond to developmental stages, environmental cues, and external stresses.
 Identification of Novel Compounds:
Identification of Novel Compounds:
Through advanced metabolomics techniques, we unearth novel and bioactive compounds within plant natural product biosynthetic pathways. This not only expands the chemical repertoire of plant-derived compounds but also presents exciting opportunities for applications in medicine, agriculture, and industry.
 Regulatory Mechanism Deciphering:
Regulatory Mechanism Deciphering:
Creative Proteomics excels in deciphering the regulatory mechanisms governing plant natural product biosynthetic pathways. Our metabolomics studies shed light on how genetic, environmental, or developmental factors influence the activation or suppression of specific biosynthetic genes, providing invaluable insights into regulatory networks.
 Optimization of Production Strategies:
Optimization of Production Strategies:
Leveraging our expertise in metabolomics, we identify critical checkpoints within biosynthetic pathways, facilitating the optimization of production strategies. This knowledge is pivotal for enhancing the yield of desired products in both plant systems and microorganisms.
 Targeted Biotechnological Interventions:
Targeted Biotechnological Interventions:
Our metabolomics data serves as a guiding framework for targeted biotechnological interventions. By manipulating key enzymes or regulatory elements identified through our research, we enable the modulation of specific natural product production. This paves the way for the development of crops with enhanced nutritional profiles and the scalable production of bioactive compounds.
 Advancements in Medicine and Drug Discovery:
Advancements in Medicine and Drug Discovery:
Creative Proteomics contributes significantly to advancements in medicine and drug discovery. Our deep understanding of plant natural product biosynthetic pathways aids in identifying potential therapeutic compounds, expediting the discovery of novel drugs, and providing crucial insights into the bioavailability and metabolism of pharmaceutical agents derived from plants.
 Holistic Systems Biology Integration:
Holistic Systems Biology Integration:
Embracing a holistic systems biology approach, we integrate metabolomics with other omics technologies. This synergy allows us to connect genetic information to the functional outputs of biosynthetic processes, providing our clients with a comprehensive understanding of plant natural product biosynthetic pathways.
Plant Natural Product Biosynthesis Metabolomics Analytical Techniques
Creative Proteomics employs state-of-the-art metabolomics techniques, with a primary focus on mass spectrometry-based methodologies. The utilization of cutting-edge instruments enhances our capability to identify, quantify, and understand the diverse metabolites involved in plant biosynthetic pathways.
High-Resolution Mass Spectrometry (HRMS): HRMS provides exceptional mass accuracy and resolution, allowing for the precise identification and quantification of a wide range of metabolites. It enhances our ability to discern subtle differences in molecular structures within complex plant metabolomes.
- Instrument: Thermo Scientific Orbitrap Fusion Lumos Tribrid Mass Spectrometer
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS facilitates the separation and analysis of metabolites with superior sensitivity. The Q Exactive Plus Mass Spectrometer, with its high-resolution capabilities, enables the detailed profiling of plant natural products, capturing intricate biosynthetic pathways.
- Instrument: Agilent 1290 Infinity II LC System coupled with Thermo Scientific Q Exactive Plus Mass Spectrometer
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is instrumental in the analysis of volatile metabolites. The Agilent 7890B GC, paired with the 5977B MSD, ensures precise separation and identification of volatile compounds involved in plant biosynthetic pathways.
- Instrument: Agilent 7890B Gas Chromatograph coupled with Agilent 5977B Mass Selective Detector (MSD)
Ultra-High-Performance Liquid Chromatography-Mass Spectrometry (UHPLC-MS): UHPLC-MS combines the efficiency of ultra-high-performance liquid chromatography with the high resolution and accurate mass measurement of the Xevo G2-XS QTof. This technology enables rapid and comprehensive profiling of plant metabolites.
- Instrument: Waters ACQUITY UPLC System coupled with Waters Xevo G2-XS QTof Mass Spectrometer
Triple Quadrupole Mass Spectrometry (QQQ-MS): QQQ-MS is employed for targeted quantification of specific metabolites. The Agilent 6460 Triple Quadrupole LC/MS system ensures high sensitivity and precision, particularly useful in studying key compounds within plant natural product biosynthetic pathways.
- Instrument: Agilent 6460 Triple Quadrupole LC/MS
Case. Metabolomic and Transcriptomic Analysis of Tanshinone Biosynthesis Induction in Salvia miltiorrhiza Hairy Root Cultures.
Background
Salvia miltiorrhiza, commonly known as Danshen, is a traditional Chinese medicinal herb known for its therapeutic properties. Tanshinones, abietane-type norditerpenoids found in Danshen, exhibit various pharmaceutical activities. Despite the medicinal significance of tanshinones, limited information exists about their biosynthesis. This study aims to elucidate the regulatory mechanisms governing tanshinone production in S. miltiorrhiza hairy root cultures.
Samples
Hairy root cultures of S. miltiorrhiza were established through Agrobacterium rhizogenes-mediated transformation. Induction was initiated using a biotic-abiotic combination of yeast extract carbohydrate fraction and Ag+. Samples were collected at various time points (0 h, 12 h, 24 h, 36 h, 48 h, 120 h, 240 h) for both transcriptome profiling and metabolite analysis.
Technological Methods
Hairy Root Culture Development and Induction:
- S. miltiorrhiza plantlets infected with Agrobacterium rhizogenes.
- Induction with a combination of yeast extract and Ag+.
Sample Collection:
- Harvested hairy roots at different time points for transcriptome profiling and metabolite analysis.
Extraction and Sample Preparation:
- Total RNA extraction using Trizol method.
- Tanshinone extraction from lyophilized hairy roots using methanol/chloroform.
UPLC-DAD-QTOF-MS Analysis:
- Metabolite analysis with UPLC-DAD-QTOF-MS using Poroshell 120 SB-C18 column.
- Gradient elution with formic acid aqueous solution and formic acid methanol solution.
- Positive ion mode mass spectrometry.
Metabolic Profile Analysis:
- LC/MS data processed using MZ-mine for integration, alignment, and explorative data analysis.
- Principal component analysis (PCA) and hierarchical clustering for metabolite profiling.
Roche 454 and Illumina GAII Sequencing:
- Reference transcriptome generated by Roche 454 FLX sequencing.
- Illumina GAII sequencing on individual samples (250–450 nt size range).
Data Analysis:
- CAP3 assembly for Roche 454 reads.
- BLASTX annotation against NCBI nr database.
- SOAP mapping for Illumina reads.
- Functional analysis, including GO, MapMan, and KEGG pathway analysis.
Transcription Factor Analysis:
- Isotigs searched against TAIR9 protein database for transcription factor annotation.
- Expression profiles clustered using k-means clustering.
Quantitative Real-Time PCR (qRT-PCR):
- qRT-PCR performed on selected genes using total RNA.
- Relative mRNA levels determined using the 2(-ΔΔC(T)) Method.
Data Availability:
- Roche 454 and Illumina GAII sequencing data deposited in NCBI Sequence Read Archive (SRA).
Results:
Metabolomic Analysis: Tanshinones showed significant increases at 120 hpi and 240 hpi, revealed by PCA.
Transcriptomic Analysis: 20,972 non-redundant genes identified, with 6,358 differentially expressed genes.
Functional Analysis: Enrichment in stress and immune response processes, down-regulation of central metabolism, and up-regulation of terpenoid biosynthesis.
Terpenoid Metabolism: Biphasic response in MVA and MEP pathways, with sustained up-regulation in tanshinone biosynthesis genes.
Co-regulated Genes: Identification of 125 cytochrome P450 genes, with eight showing potential involvement in tanshinone biosynthesis.
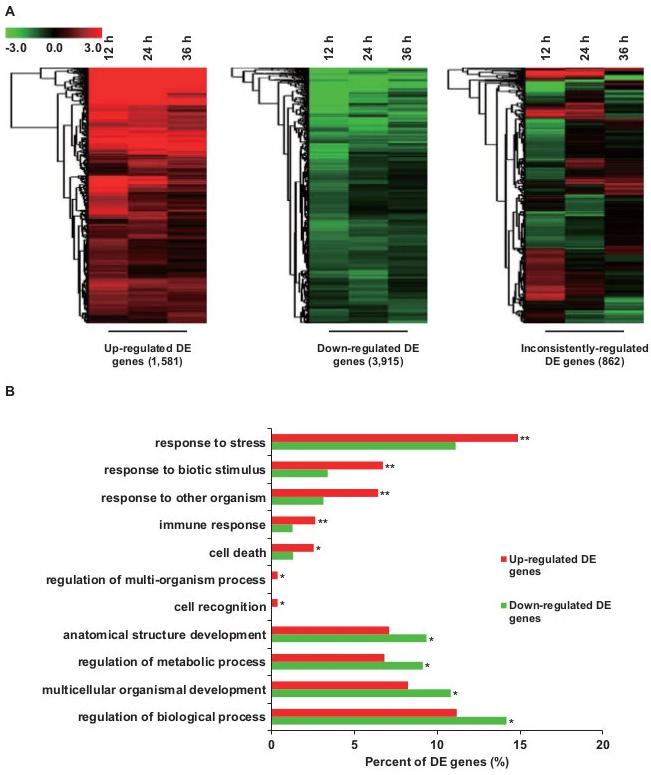 Functional analysis of differentially expressed (DE) genes.
Functional analysis of differentially expressed (DE) genes.
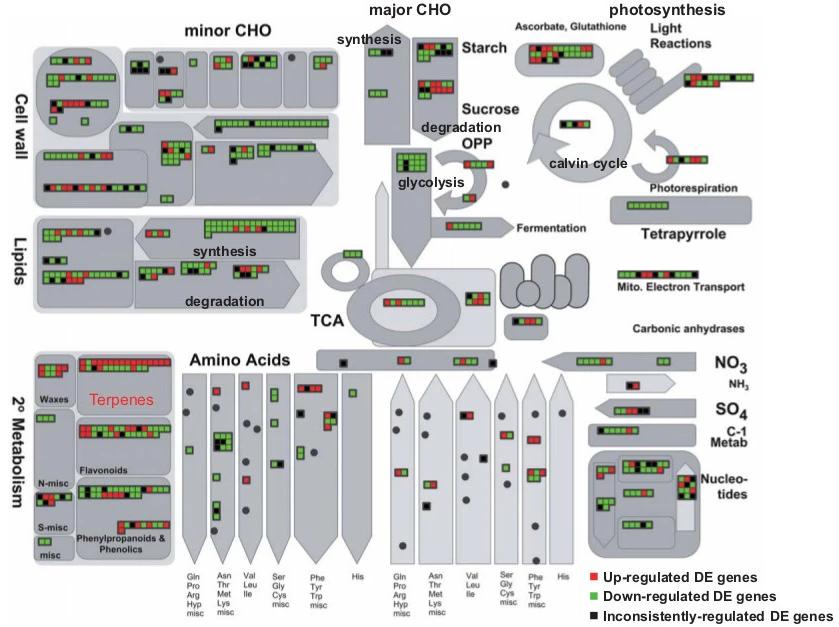 Overview of induced transcriptional changes for the enzymatic genes from various metabolic pathways.
Overview of induced transcriptional changes for the enzymatic genes from various metabolic pathways.
Reference
- Gao, Wei, et al. "Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza." BMC genomics 15 (2014): 1-14.


 Functional analysis of differentially expressed (DE) genes.
Functional analysis of differentially expressed (DE) genes. Overview of induced transcriptional changes for the enzymatic genes from various metabolic pathways.
Overview of induced transcriptional changes for the enzymatic genes from various metabolic pathways.