Why Metabolomics for Forestry Research?
Comprehensive Insight into Tree Metabolism
Metabolomics offers a comprehensive snapshot of the metabolic landscape within trees. Unlike genomics or transcriptomics, which focus on genes and their expression, metabolomics provides a real-time portrayal of the biochemical activity within a tree. This holistic view enables researchers to grasp how trees respond to changing environmental conditions, diseases, and external factors.
Dynamic Responses to Environmental Changes
Forests, as dynamic ecosystems, are subjected to an array of environmental changes. Metabolomics facilitates the observation of dynamic responses at the molecular level. Identifying and quantifying specific metabolites grants insights into the mechanisms underlying stress responses, resilience, and overall health of forest ecosystems.
Identifying Biomarkers for Tree Health
A pivotal aspect of metabolomics in forestry is the discovery of biomarkers indicative of tree health. These biomarkers act as molecular signatures, offering early indicators of stress or disease. The ability to identify and monitor these molecular cues enhances our capacity to diagnose and mitigate threats to forest ecosystems.
With a commitment to precision and grounded scientific exploration, we offer tailored solutions to unravel the metabolic dynamics within forest ecosystems. Our forestry metabolomics services address a spectrum of needs, from profiling diverse tree species to analyzing stress responses and contributing to sustainable forestry practices. We utilize advanced mass spectrometry-based techniques, including Liquid Chromatography-Mass Spectrometry (LC-MS) and Gas Chromatography-Mass Spectrometry (GC-MS) for accurate and sensitive analysis.
Beyond exploration, our services aim to contribute practical insights to sustainable forestry practices. We delve into metabolic pathways, aiming to guide forest management strategies that balance conservation and human needs.
Forestry Metabolomics Solutions
Explore the intricate world of forestry with Creative Proteomics' advanced Metabolomics research services. Our specialized offerings cater to diverse research directions, contributing to a deeper understanding of tree species, stress responses, sustainable practices, biogeography, and overall forest health.
 Metabolic Profiling of Tree Species
Metabolic Profiling of Tree Species
Delve into the unique metabolic signatures of various tree species. Our advanced metabolomic profiling provides tailored insights into conifers and deciduous varieties, unraveling specific biochemical nuances.
 Stress Response Metabolomics
Stress Response Metabolomics
Navigate the dynamic stressors impacting forests. From climate challenges to pollutants and pathogens, our stress response metabolomics projects map adaptive strategies, offering targeted insights for enhancing forest resilience.
 Metabolomics in Sustainable Forestry Practices
Metabolomics in Sustainable Forestry Practices
Contribute to sustainable forest management. Our metabolomics projects optimize resource utilization, guiding forest management strategies that harmonize conservation and human needs.
 Biogeography and Metabolomics
Biogeography and Metabolomics
Unravel the influence of geography on tree metabolism. Our research explores how geographic factors shape metabolic profiles, providing insights into variation across different regions.
 Forest Health Monitoring through Metabolomics
Forest Health Monitoring through Metabolomics
Implement proactive measures for forest health. Our metabolomics research establishes baseline profiles and monitors changes, enabling early detection of disturbances, diseases, and environmental threats.
Forestry Metabolomics Analytical Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS)
Our high-performance LC-MS system, the Thermo Scientific Q Exactive HF-X, sets the standard for metabolite analysis. This state-of-the-art instrument ensures accurate and sensitive detection across a broad range of metabolites, enhancing resolution and precision. The combination of liquid chromatography and mass spectrometry allows for in-depth profiling of complex biological samples, providing detailed insights into the metabolic dynamics of trees.
Gas Chromatography-Mass Spectrometry (GC-MS)
For the analysis of volatile and semi-volatile metabolites, Creative Proteomics relies on the Agilent 7890B/5977A GC-MSD system. This advanced technology facilitates the separation and characterization of compounds with high sensitivity. By capturing the unique signatures of volatile metabolites, we contribute to a comprehensive understanding of the ecological interactions and emission patterns of trees.
Nuclear Magnetic Resonance (NMR) Spectroscopy
Harnessing the power of high-field NMR spectroscopy, our Bruker Avance III 600 MHz provides invaluable insights into the structural details of metabolites. This non-destructive technique allows for the elucidation of molecular structures, enhancing our ability to comprehensively profile and identify key metabolites within tree samples.
High-Resolution Mass Spectrometry (HRMS)
Creative Proteomics utilizes the Thermo Scientific Orbitrap Eclipse Tribrid Mass Spectrometer for high-resolution mass spectrometry (HRMS). This instrument ensures unparalleled accuracy in identifying and quantifying metabolites. The high resolution and mass accuracy of this technology enable the precise characterization of complex metabolomic profiles, contributing to a deeper understanding of the metabolic dynamics within forestry systems.
Forestry Metabolomics Data Analysis
| Data Analysis Step |
Description |
| Preprocessing |
- Raw data preprocessing to remove noise and artifacts.
- Peak detection and alignment for accurate metabolite identification. |
| Feature Extraction |
- Extraction of relevant features, including m/z values, retention times, and peak intensities.
- Ensures comprehensive coverage of metabolomic profiles. |
| Metabolite Identification |
- Utilization of spectral databases and reference libraries for accurate metabolite identification.
- Confirmation of identified metabolites through MS/MS spectra. |
| Quantification |
- Integration of peak areas for accurate metabolite quantification.
- Calibration using internal or external standards for concentration determination. |
| Statistical Analysis |
- Application of statistical methods (e.g., t-tests, ANOVA, PCA) to identify significant metabolomic changes.
- Clustering analysis for pattern recognition. |
| Pathway Analysis |
- Mapping identified metabolites onto metabolic pathways.
- Identification of key pathways associated with specific biological processes. |
| Visualization |
- Generation of visual representations (e.g., heatmaps, volcano plots) for intuitive interpretation of complex data.
- Interactive data exploration tools for enhanced understanding. |
Case. Comparative Metabolomics Reveals Contrasting Chemical Diversity and Phylogenetic Signals in Tropical and Temperate Plant Communities
Background
The study investigates the role of chemical interactions in shaping plant communities, focusing on the divergence of leaf metabolomes in tropical (Barro Colorado Island, BCI) and temperate (Smithsonian Environmental Research Center, SERC) forests. It explores the potential influence of plant-enemy interactions on chemical diversity and its relationship with phylogenetic signals.
Samples
BCI (Tropical): Sampled 138 species, including the 48 most abundant species and representatives from seven species-rich genera.
SERC (Temperate): Sampled 18 invasive and 47 native species, including all species in the three most species-rich genera.
Technological Methods
- Leaf Collection and Analysis: Expanding, unlignified leaves collected from shaded understory. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) employed for chemical analysis.
- Chemical Structural-Compositional Similarity (CSCS): Developed a metric to quantify chemical structural-compositional similarity over all compounds in two species. Used phylogenetically independent contrasts (PICs) to test relationships between chemical similarity and phylogenetic distance.
- Phylogenetic Signal Analysis: Regression of CSCSmrca against log-transformed phylogenetic distance for evaluating phylogenetic signal.
- Statistical Analyses: One-way ANOVA, phylogenetic ANOVA, and non-metric multidimensional scaling (NMDS) utilized to compare chemical spaces and assess differences between tropical and temperate communities.
Results:
Phylogenetic Signal Contrasts:
- BCI (Tropical): Lack of phylogenetic signal in leaf metabolomes, suggesting rapid chemical divergence among closely related species, possibly driven by selection or competitive exclusion.
- SERC (Temperate): Clear phylogenetic signal in leaf metabolomes, indicating similarity among closely related species, contrary to the hypothesis of strong selection for divergence.
Chemical Differences Between Tropical and Temperate Trees:
- Fundamental chemical differences observed between tropical and temperate tree species, supporting the hypothesis of stronger plant-enemy interactions in the tropics.
- Tropical species exhibit greater qualitative chemical differences, potentially driven by selection for divergence in secondary metabolites.
Contrast with Leaf Functional Traits:
- Leaf metabolomes diverge more rapidly than leaf functional traits in tropical trees after speciation, supporting the idea that traits with a greater potential for reciprocal coevolution between plants and their enemies promote diversification, especially in low latitudes.
Global Latitudinal Diversity Gradient:
- Controversy surrounding the hypothesis of more intense biotic interactions in the tropics is discussed.
- Qualitative differences in chemical defenses are suggested as a factor influencing the global latitudinal diversity gradient, with tropical species exhibiting greater novelty in defense.
Implications and Future Directions:
- The study highlights the need for collaborative efforts to integrate metabolomics with ecological factors across diverse sites and latitudinal gradients.
- Qualitative differences in chemical defenses may play a crucial role in niche partitioning and lineage diversification at global ecological scales.
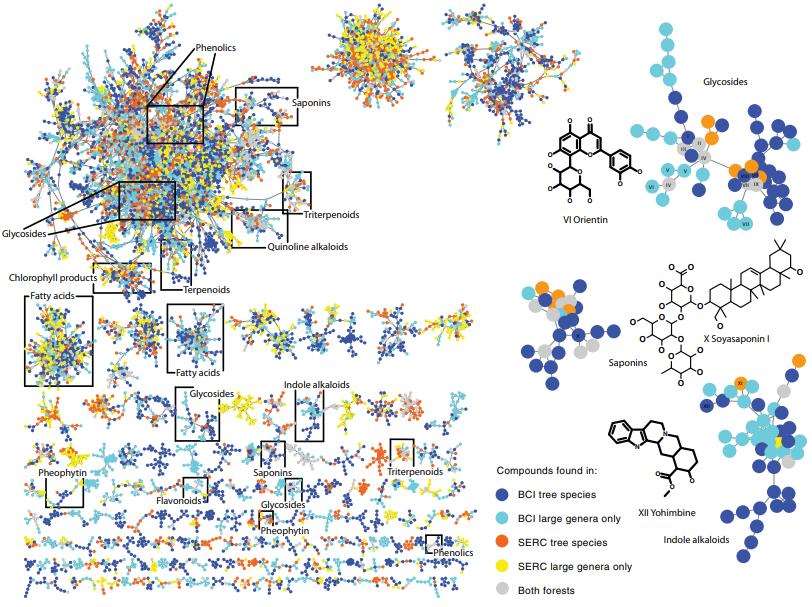 Molecular network of 36,223 compounds in leaves of tree and shrub species from SERC, Maryland and BCI, Panama.
Molecular network of 36,223 compounds in leaves of tree and shrub species from SERC, Maryland and BCI, Panama.
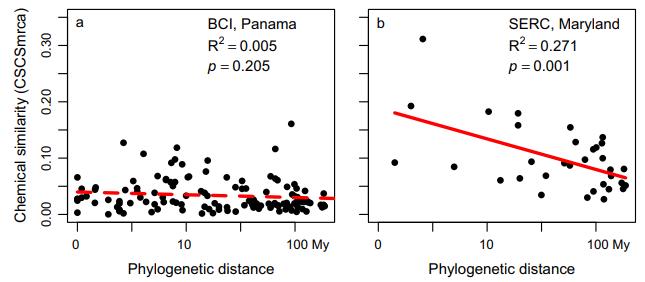 Relationships between the mean chemical similarity of species descended from each node (or most recent common ancestor, CSCSmrca) and log-transformed phylogenetic distance for BCI (panel a) and SERC (b).
Relationships between the mean chemical similarity of species descended from each node (or most recent common ancestor, CSCSmrca) and log-transformed phylogenetic distance for BCI (panel a) and SERC (b).
Reference
- Sedio, Brian E., et al. "Comparative foliar metabolomics of a tropical and a temperate forest community." (2018): 2647-2653.


 Molecular network of 36,223 compounds in leaves of tree and shrub species from SERC, Maryland and BCI, Panama.
Molecular network of 36,223 compounds in leaves of tree and shrub species from SERC, Maryland and BCI, Panama. Relationships between the mean chemical similarity of species descended from each node (or most recent common ancestor, CSCSmrca) and log-transformed phylogenetic distance for BCI (panel a) and SERC (b).
Relationships between the mean chemical similarity of species descended from each node (or most recent common ancestor, CSCSmrca) and log-transformed phylogenetic distance for BCI (panel a) and SERC (b).