What is Gossypium (Cotton)?
Gossypium, commonly known as cotton, is a plant renowned for its valuable natural fibers. Metabolomics, the study of its metabolic processes, is crucial for understanding its biology, enhancing fiber quality, addressing agricultural challenges, promoting sustainability, and advancing biotechnology in cotton farming. This comprehensive approach to research provides insights into the plant's growth, development, and responses to various environmental factors, contributing to the improvement of cotton crops and the textile industry as a whole.
Our Gossypium (Cotton) Metabolomics Analysis Projects
Metabolic Profiling: We conduct comprehensive metabolic profiling of cotton plants to identify and quantify a broad spectrum of metabolites. This analysis helps researchers gain insights into the plant's primary and secondary metabolism, including the synthesis of essential compounds, energy production, and responses to environmental stresses.
Stress Response Mechanisms: Metabolomics is instrumental in unraveling how cotton plants respond to various stresses such as drought, pests, and diseases. By studying metabolic changes, we can identify specific pathways and metabolites involved in stress adaptation and resilience, which can inform strategies for crop protection and improvement.
Fiber Development: Cotton is primarily cultivated for its fibers. We use metabolomics to investigate the metabolic pathways associated with fiber development. This includes studying the biosynthesis of cellulose, hemicellulose, and lignin, which are critical components of cotton fibers. Understanding these pathways aids in enhancing fiber quality and yield.
Nutrient Uptake and Utilization: Metabolomics helps us examine how cotton plants acquire, transport, and utilize essential nutrients such as nitrogen, phosphorus, and potassium. This knowledge is pivotal for optimizing nutrient management in cotton farming to ensure healthy crop growth and maximum yield.
Secondary Metabolite Profiling: Cotton plants produce secondary metabolites with various ecological and industrial significance. Metabolomic analysis allows us to identify and quantify these compounds, including phytochemicals with potential applications in pharmaceuticals, textiles, and other industries.
Genetic Engineering: In the pursuit of genetically improved cotton varieties, we apply metabolomics to assess the impact of genetic modifications on the plant's metabolic pathways. This evaluation helps in selecting and fine-tuning traits, such as pest resistance or drought tolerance.
Sustainable Agriculture: Metabolomics aids in the development of sustainable agricultural practices for cotton farming. By understanding the plant's metabolic responses to different cultivation techniques, we can optimize resource utilization and reduce environmental impacts.
Biotechnological Applications: We leverage metabolomics to support biotechnological advancements in cotton research. This includes the characterization of genetically modified cotton plants and assessing the safety and efficacy of biotech interventions.
Analytical Techniques for Gossypium (Cotton) Metabolomics
1. Sample Preparation: The process typically begins with sample collection and preparation. Cotton plant tissues, such as leaves, stems, or fibers, are harvested, freeze-dried, and ground into a fine powder to facilitate metabolite extraction.
2. Metabolite Extraction: Metabolites are extracted from the powdered samples using appropriate solvents. Commonly used solvents include methanol, ethanol, and water, sometimes with the addition of internal standards for quantification.
3. Sample Analysis by Mass Spectrometry:
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS is a widely used technique for metabolomics analysis. High-performance liquid chromatography (HPLC) is coupled with mass spectrometry to separate and identify metabolites. Common LC-MS instruments include:
- Agilent 1290 Infinity II LC System coupled with an Agilent 6545 Q-TOF Mass Spectrometer.
- Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer.
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is another powerful technique, especially for volatile and thermally stable metabolites. Cotton samples are derivatized before analysis. Common GC-MS instruments include:
- Agilent 7890B GC System coupled with an Agilent 5977B Mass Spectrometer.
- Shimadzu GCMS-QP2020 NX Gas Chromatograph-Mass Spectrometer.
4. Data Acquisition and Analysis:
- The mass spectrometers generate data in the form of mass spectra, which are then processed and analyzed using specialized software. Common software tools include XCMS, MetaboAnalyst, and MZmine.
- Metabolites are identified by comparing mass spectra and retention times with reference libraries and databases.
5. Metabolite Annotation: The identified metabolites are annotated based on their chemical properties, such as mass-to-charge ratio (m/z), retention time, and fragmentation patterns. This step is critical for assigning biological significance to the detected metabolites.
6. Statistical Analysis: Statistical methods, such as multivariate analysis and pathway analysis, are applied to identify significant changes in metabolite levels under different experimental conditions. This helps in understanding the metabolic responses of cotton plants.
7. Integration with Genomics and Proteomics: Metabolomics data can be integrated with genomics and proteomics data to gain a comprehensive understanding of how genes, proteins, and metabolites interact in cotton plants at the protein level.
8. Biological Interpretation: The results of metabolomics analysis provide insights into various aspects of cotton metabolism, including primary and secondary metabolism, stress responses, and the production of valuable compounds like cellulose in cotton fibers.
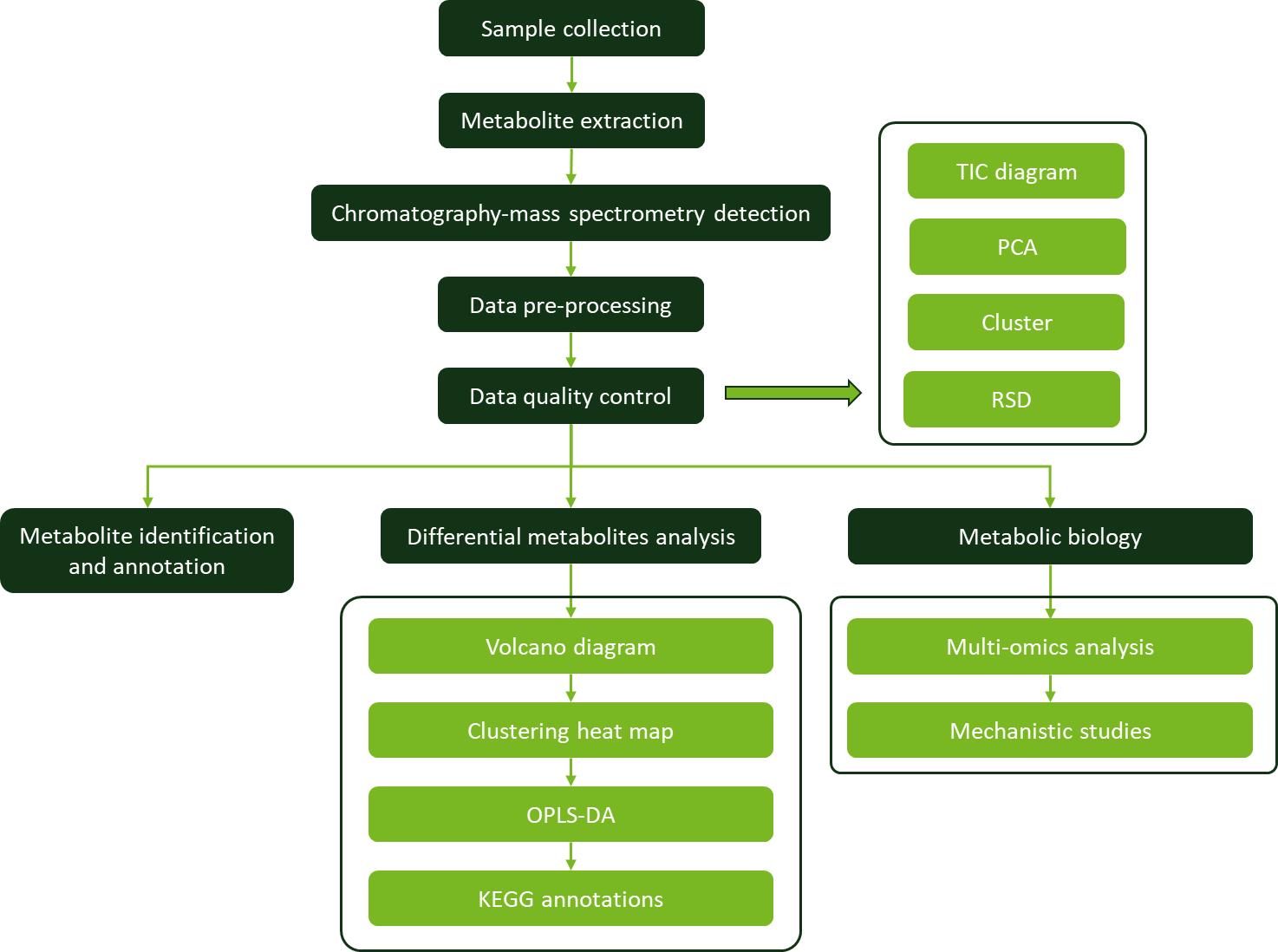 Workflow for Metabolomics Service
Workflow for Metabolomics Service
Sample Requirements for Cotton Metabolomics
| Sample Type |
Tissue |
Sample Size |
Storage |
Notes |
| Leaves |
Fresh or Frozen |
Approx. 1-2 g |
-80°C in Foil |
Harvested at specific growth stage |
| Stems |
Fresh or Frozen |
Approx. 1-2 g |
-80°C in Foil |
Harvested at specific growth stage |
| Cotton Fibers |
Fresh or Dried |
Approx. 0.5-1 g |
Room Temp. in Bags |
Separate by developmental stage |
| Roots |
Fresh or Frozen |
Approx. 1-2 g |
-80°C in Foil |
Harvested at specific growth stage |
| Soil (Rhizosphere) |
N/A |
Soil Sample |
-20°C |
Collect around cotton plant roots |
Case. Molecular and Biochemical Strategies for Enhancing Cotton Fiber Length
Background
Cotton fiber length is a crucial trait for the textile industry, and improving it has been a goal in cotton breeding and biotechnology. This study investigates molecular and biochemical mechanisms to increase cotton fiber length using transcriptomic and metabolomic data.
Samples
Two cotton cultivars were used in this study: Gossypium hirsutum cv Deltapine 90 (Gh) and Gossypium barbadense cv Phytogen 800 (Gb). Fiber samples were collected at various days post-anthesis (DPA) to capture different stages of fiber development.
Technological Methods
- RNA Extraction and Sequencing: RNA was extracted from cotton fiber samples and sequenced using Illumina technology. Libraries were prepared for each sample, and sequencing was performed to generate a comprehensive transcriptome dataset.
- Mapping and Annotation: A reference set was created by combining genomic data from both the D genome (G. raimondii - Gr) and the A genome (G. arboreum - Ga). This reference was used to map and annotate the sequenced transcripts. Gene expression levels were quantified using Reads Per Kilobase Million (RPKM).
- Differential Gene Expression Analysis: Pairwise comparisons were made within and between cotton genotypes (Gh and Gb) and DPA stages to identify differentially expressed genes. Statistical tests and multiple correction methods were employed to determine significant gene expression changes.
- Metabolomics Analysis: Metabolites in cotton fiber samples were analyzed using Liquid Chromatography/Mass Spectrometry (LC/MS) and Gas Chromatography/Mass Spectrometry (GC/MS). Data normalization and imputation of missing values were performed.
- Data Analysis and Visualization: Bioinformatics tools and R programming were used for overrepresentation analysis of KEGG pathways, cluster analysis, and generation of heat maps and plots. Venn diagrams were created to identify overlapping gene sets.
Results
Transcriptomic and metabolomic data indicated the potential to enhance cotton fiber length through strategies that increase ascorbate levels for ROS scavenging and recycling.
Over-expression of specific genes, such as chloroplast-targeted APX, showed promising preliminary results, with a 10% increase in Gh fiber length.
Comparative analysis of gene sets revealed specific transcripts associated with continued fiber elongation in Gb fiber, including genes related to water transport, pectin degradation, and glycosylation of flavonoids.
Metabolomic data highlighted differences in metabolite concentrations between Gb and Gh fibers, emphasizing the role of metabolites like ascorbate, flavonoids, and oligosaccharides in managing ROS and supporting prolonged fiber elongation.
The study provided valuable insights into the genetic and biochemical factors influencing cotton fiber development and suggested avenues for further research and potential strategies for cotton breeding and biotechnology to enhance fiber length.
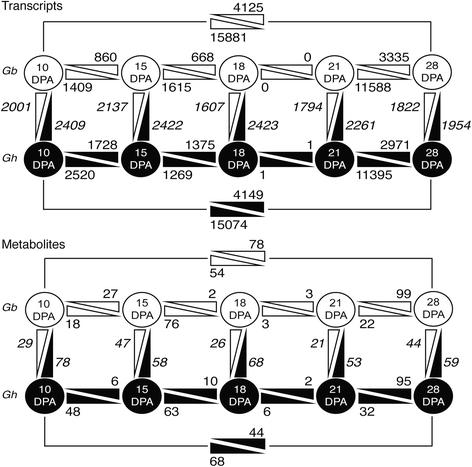 Quantitative changes in the transcriptome and metabolome within and between Gb and Gh fibers.
Quantitative changes in the transcriptome and metabolome within and between Gb and Gh fibers.
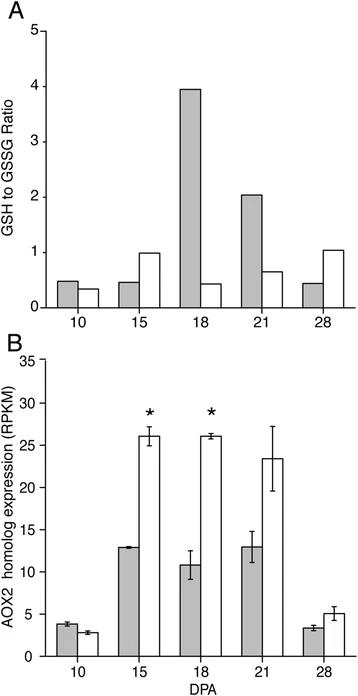 Metabolomic and transcriptomic evidence that transition-stage Gh fiber experiences more oxidative stress than Gb fiber.
Metabolomic and transcriptomic evidence that transition-stage Gh fiber experiences more oxidative stress than Gb fiber.
Reference
- Tuttle, John R., et al. "Metabolomic and transcriptomic insights into how cotton fiber transitions to secondary wall synthesis, represses lignification, and prolongs elongation." BMC genomics 16.1 (2015): 1-28.


 Workflow for Metabolomics Service
Workflow for Metabolomics Service Quantitative changes in the transcriptome and metabolome within and between Gb and Gh fibers.
Quantitative changes in the transcriptome and metabolome within and between Gb and Gh fibers. Metabolomic and transcriptomic evidence that transition-stage Gh fiber experiences more oxidative stress than Gb fiber.
Metabolomic and transcriptomic evidence that transition-stage Gh fiber experiences more oxidative stress than Gb fiber.