What is Olea europaea (Olive Tree)?
Olea europaea, commonly known as the Olive tree, is a species of small evergreen tree native to the Mediterranean Basin and parts of Asia and Africa. It belongs to the Oleaceae family and is renowned for its economic and cultural significance, primarily due to the production of olives and olive oil.
Olive trees have distinctive silvery-green, lance-shaped leaves and gnarled trunks that can live for centuries, making them a symbol of longevity and strength. They produce small, cream-colored flowers in spring, which give rise to olives in various shapes and sizes, depending on the variety. Olives are harvested for both culinary and industrial purposes, with their most prized product being olive oil, a key component of Mediterranean cuisine and a staple of many international dishes.
Metabolic analysis of the Olive tree involves the study of its metabolic processes, including the chemical reactions and pathways that occur within the plant's cells to maintain life and produce important compounds. This analysis is crucial for understanding the growth, development, and adaptation of Olive trees to various environmental conditions. Olive tree metabolic analysis allows you to understand the following:
- Primary Metabolism: This includes the basic metabolic pathways responsible for essential functions like photosynthesis, respiration, and nutrient assimilation. Olive trees, like other plants, utilize photosynthesis to convert sunlight, water, and carbon dioxide into energy (in the form of sugars) and oxygen.
- Secondary Metabolism: Olive trees are known for producing a range of secondary metabolites, such as phenolic compounds, terpenes, and flavonoids. These compounds contribute to the tree's defense mechanisms, flavor and aroma profiles of olives and olive oil, and potential health benefits.
- Olive Oil Biosynthesis: Metabolic analysis of Olive trees often focuses on the biosynthesis of olive oil within the fruit. This process involves the conversion of fatty acids and glycerol into triglycerides, which accumulate in olive fruit cells. Understanding the biochemical pathways behind olive oil production is vital for optimizing olive cultivation and oil quality.
- Response to Environmental Stress: Olive trees are known for their ability to thrive in challenging environmental conditions, such as drought and salinity. Metabolic analysis helps uncover the molecular mechanisms that allow Olive trees to adapt and survive under stress, offering insights for crop improvement.
- Genetic and Metabolic Diversity: Different Olive tree varieties exhibit variations in their metabolic profiles and nutritional content. Researchers use metabolic analysis to identify and characterize these differences, aiding in the selection of varieties with desirable traits.
Specific Olive Analysis Projects by Creative Proteomics
Olive Oil Quality Assessment
Project Description: Olive oil is a prized culinary commodity with a rich history and cultural significance. Ensuring its quality and authenticity is of paramount importance. Our Olive Oil Quality Assessment project leverages advanced metabolomics techniques to scrutinize the composition of olive oil, providing precise insights into its purity, freshness, and potential adulteration.
Key Objectives:
- Identification of adulterants or contaminants in olive oil.
- Quantification of essential compounds responsible for taste, aroma, and health benefits.
- Validation of olive oil quality labels for producers and consumers.
Environmental Stress Response
Project Description: Olive Trees are renowned for their resilience to the Mediterranean climate, which can be characterized by harsh conditions such as drought, extreme temperatures, and soil salinity. Our Environmental Stress Response project investigates how Olive Trees adapt to and mitigate the effects of these environmental stressors at the metabolic level.
Key Objectives:
- Identification of metabolic pathways involved in stress response.
- Discovery of stress-specific biomarkers.
- Development of strategies for enhancing Olive Tree resilience in changing climates.
Nutraceutical Development
Project Description: Olive Trees produce a diverse array of phytochemicals, some of which have demonstrated significant health-promoting properties. Our Nutraceutical Development project focuses on the identification and quantification of these bioactive compounds, laying the foundation for the development of nutraceuticals and functional foods.
Key Objectives:
- Comprehensive profiling of health-beneficial phytochemicals.
- Assessment of their bioavailability and efficacy.
- Support for the creation of health-enhancing products.
Phytochemical Profiling
Project Description: The phytochemical diversity of Olive Trees extends beyond olive oil, encompassing various parts of the tree, including leaves and fruit. Our Phytochemical Profiling project employs metabolomics analysis to uncover the full spectrum of phytochemicals present in different tree components.
Key Objectives:
- Identification and quantification of phytochemicals with potential industrial and health applications.
- Selection and breeding of Olive Tree cultivars enriched in specific phytochemicals.
- Tailored strategies for maximizing phytochemical yield.
Olive Metabolomics Analysis Techniques
| Analytical Technique |
Instrumentation |
Description |
| Liquid Chromatography-Mass Spectrometry (LC-MS) |
Thermo Scientific™ Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer |
High-resolution, sensitive instrument combining quadrupole mass filter with Orbitrap mass analyzer; compatible with positive and negative ionization modes. |
| Gas Chromatography-Mass Spectrometry (GC-MS) |
Agilent 7890B Gas Chromatograph coupled to an Agilent 5977A MSD |
Robust system for analyzing volatile and thermally stable metabolites; precise temperature control, high-performance mass selective detection. |
| Nuclear Magnetic Resonance (NMR) Spectroscopy |
Bruker Avance III HD NMR Spectrometer |
Non-destructive technique for structural and quantitative analysis; high-resolution capabilities, detects multiple nuclei. |
| Ultra-High Performance Liquid Chromatography-Mass Spectrometry (UHPLC-MS) |
Waters ACQUITY UPLC coupled to a Waters Xevo G2-XS QToF Mass Spectrometer |
Rapid separation and characterization of metabolites; features ultra-high performance liquid chromatography, QToF mass spectrometry, and data-independent acquisition (DIA). |
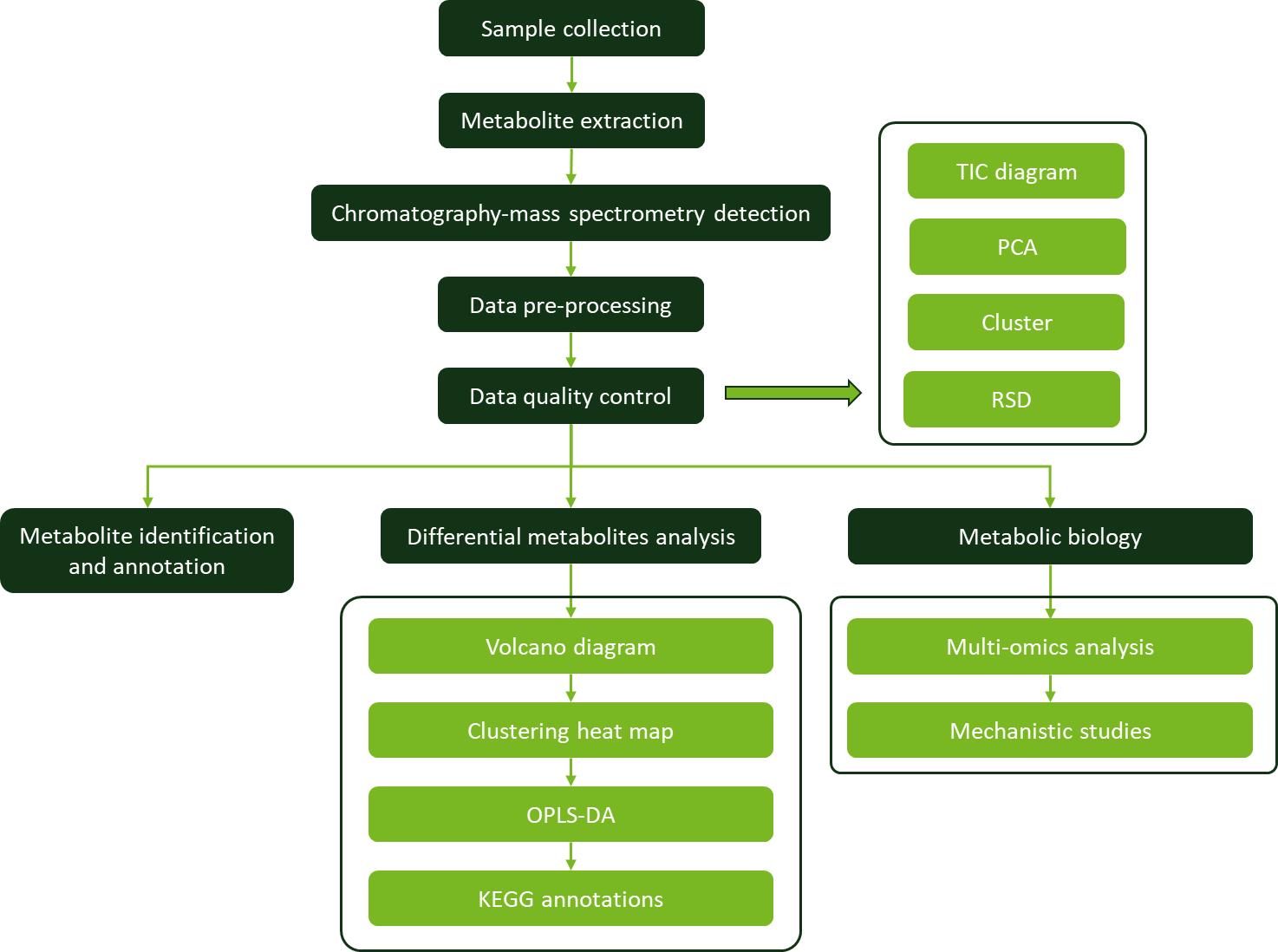 Workflow for Metabolomics Service
Workflow for Metabolomics Service
Sample Requirements for Olive Tree Metabolomics
| Sample Type |
Sample Size |
| Leaves |
100g - 200g |
| Fruit |
100g - 200g |
| Roots |
50g - 100g |
| Xylem Sap |
5ml - 10ml |
| Flowers |
Varies (as needed) |
| Bark |
Varies (as needed) |
| Trunk |
Varies (as needed) |
| Seeds |
Varies (as needed) |
| Soil Near Roots |
Varies (as needed) |
Case. Identification of Molecular Markers for Olive Quick Decline Syndrome (OQDS) Using Metabolomics
Background
The study focuses on OQDS, a devastating disease affecting olive trees in the Salento Peninsula, Italy, caused by Xylella fastidiosa (Xf) infection. OQDS leads to the desiccation and death of olive trees and has significant economic implications. Xf is a Gram-negative pathogen that obstructs water and mineral salt uptake in plants' apical regions. This study aims to identify molecular markers to distinguish between healthy and infected olive trees, providing a potential tool for early disease detection.
Samples
The study encompasses four groups of plant extracts: "Puglia infected," "Liguria healthy," "Puglia healthy," and "Puglia desiccated." These samples represent different states (healthy, infected, and desiccated) and regions (Puglia and Liguria) of olive trees.
Technological Methods
HPLC-HRMS Analytical Method Development: The analytical method involves a multi-step gradient HPLC separation, optimized to separate a wide range of metabolites, from organic acids to phytosterols. Mass spectrometry (MS) is performed in the negative ion mode for improved sensitivity and reduced background noise.
Sample Two-Dimensional Clustering: Principal component analysis (PCA) is employed to cluster samples based on their metabolite profiles. The study identifies three main clusters: quality control (QC) samples, infected samples, and healthy/desiccated samples. It also explores the potential influence of regional differences.
Features Annotation: Metabolites are annotated using tandem MS experiments and databases like MoNA, UNPD, and MetFrag. Several annotated compounds, including plant secondary metabolites like jasmonate sulfate and coumarins, are found to play roles in plant defense and stress responses.
Feature Abundance Class-Related Variability: Abundance boxplots reveal that most annotated features are up-regulated in infected samples, with some variability related to regionality. This suggests that infection and regionality influence the metabolite profiles of olive trees.
Results
The study successfully identifies molecular markers that differentiate between healthy and OQDS-infected olive trees. These markers, including jasmonate sulfate, coumarins, and phytosteroids, highlight the plant's defense and stress responses. The research provides a foundation for the development of targeted analytical methods for early OQDS detection and offers insights into the metabolome of olive trees under infection stress.
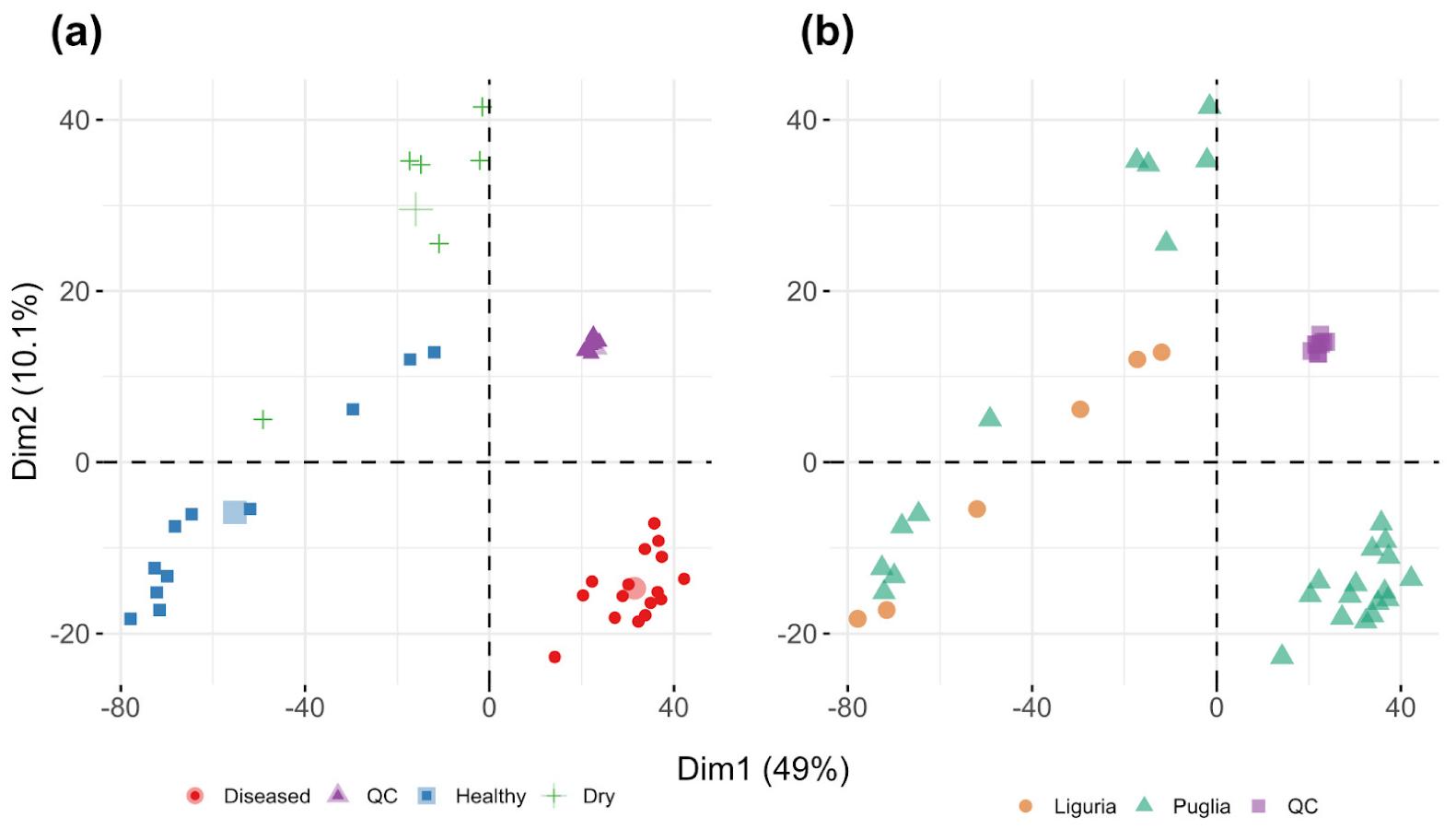 PCA score plots for all the samples analyzed
PCA score plots for all the samples analyzed
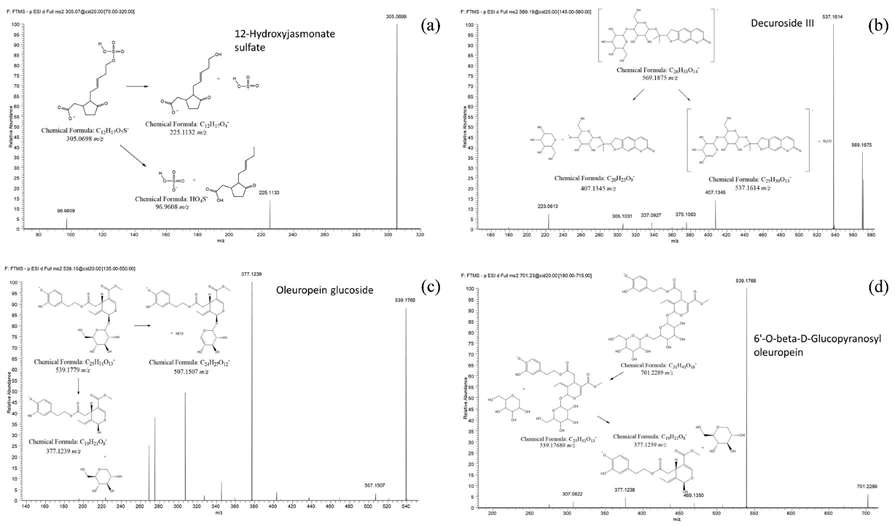 MS2 spectra of some annotated features together with their fragmentation patterns: 12-Hydroxyjasmonate sulfate (a), Decuroside III (b), Oleuropein glucoside (c) and 6'-O-beta-D-Glucopyranosyl-oleuropein (d)
MS2 spectra of some annotated features together with their fragmentation patterns: 12-Hydroxyjasmonate sulfate (a), Decuroside III (b), Oleuropein glucoside (c) and 6'-O-beta-D-Glucopyranosyl-oleuropein (d)
Reference
- Asteggiano, Alberto, et al. "HPLC-HRMS global metabolomics approach for the diagnosis of "Olive Quick Decline Syndrome" markers in olive trees leaves." Metabolites 11.1 (2021): 40.


 Workflow for Metabolomics Service
Workflow for Metabolomics Service PCA score plots for all the samples analyzed
PCA score plots for all the samples analyzed MS2 spectra of some annotated features together with their fragmentation patterns: 12-Hydroxyjasmonate sulfate (a), Decuroside III (b), Oleuropein glucoside (c) and 6'-O-beta-D-Glucopyranosyl-oleuropein (d)
MS2 spectra of some annotated features together with their fragmentation patterns: 12-Hydroxyjasmonate sulfate (a), Decuroside III (b), Oleuropein glucoside (c) and 6'-O-beta-D-Glucopyranosyl-oleuropein (d)