What is Helianthus annuus (Common Sunflower)?
Helianthus annuus, commonly known as the "Common Sunflower," is a well-known and widely cultivated plant species. It belongs to the genus "Helianthus" and is part of the family Asteraceae. This plant is native to North America, specifically regions of the United States and Mexico, but it is now grown in many parts of the world for various purposes. The Common Sunflower is recognized for its characteristic large, bright yellow flower heads and tall stems.
Metabolomics is a field of science that involves the comprehensive study of small molecules, known as metabolites, within an organism, tissue, or cell at a specific point in time. In the context of Helianthus annuus (Common Sunflower), metabolomics analysis refers to the systematic and detailed examination of the complete set of metabolites present in the plant.
Specific Common Sunflower Analysis Projects by Creative Proteomics
At Creative Proteomics, we offer specialized services for Helianthus annuus metabolomics analysis to support research endeavors in the following areas:
Metabolite Profiling: Perform a comprehensive analysis of primary and secondary metabolites, including carbohydrates, amino acids, lipids, organic acids, and secondary compounds, to establish a comprehensive metabolic profile of Helianthus annuus.
Stress Responses: Investigate the dynamic metabolic responses of sunflowers to a broad spectrum of environmental stresses, such as drought, salinity, temperature variations, pathogen infections, and herbivore attacks, aiming to elucidate stress-induced metabolic alterations and adaptive strategies.
Nutrient Utilization and Allocation: Study nutrient uptake, transport, and allocation within the plant, with a focus on understanding metabolic processes governing nutrient utilization (e.g., nitrogen, phosphorus, and micronutrients) to optimize nutrient management practices for enhanced sunflower growth and yield.
Secondary Metabolite Production: Identify, quantify, and characterize the key secondary metabolites, including terpenoids, phenolic compounds, and alkaloids, responsible for sunflower's pharmaceutical, cosmetic, and industrial applications. Explore metabolic engineering strategies to boost secondary metabolite production.
Metabolic Regulatory Networks: Investigate the regulatory networks controlling sunflower metabolism, including transcription factors, enzymes, and signaling pathways, to uncover key regulatory nodes that can be targeted for metabolic engineering and crop improvement.
Developmental Metabolomics: Examine the dynamic changes in metabolic profiles across different developmental stages of Helianthus annuus, from germination and vegetative growth to flowering and seed development, to gain insights into the evolution of metabolic pathways during the plant's life cycle.
Metabolic Signatures for Stress Resilience: Identify unique metabolic signatures associated with stress resistance and susceptibility, allowing for the development of biomarkers that aid in the selection and breeding of stress-tolerant sunflower cultivars.
Nutritional Quality Enhancement: Assess the nutritional composition of sunflower seeds, oils, and other edible parts by analyzing essential nutrients, antioxidants, and anti-nutritional factors, contributing to the development of sunflower varieties with improved nutritional profiles.
Metabolomics-Guided Breeding: Utilize metabolomics data to guide breeding programs aimed at developing sunflower varieties with desired metabolic traits, such as enhanced oil content, altered fatty acid composition, or increased resistance to specific pests or diseases.
Integration with Genomics and Transcriptomics: Integrate metabolomics data with genomic and transcriptomic datasets to establish comprehensive multi-omics networks, facilitating a systems biology approach to understanding Helianthus annuus metabolism.
Biofuel Production: Investigate metabolic pathways related to lipid metabolism and cellulose breakdown, providing insights into sunflower's potential as a biofuel feedstock and informing strategies for bioenergy production.
Environmental Adaptation: Study how Helianthus annuus adapts its metabolism to different ecological niches, including wild and domesticated varieties, to gain insights into natural variation and domestication processes.
Pharmacological Applications: Explore the metabolic basis for sunflower's traditional medicinal uses and investigate its potential in modern pharmaceutical applications.
Common Sunflower Metabolomics Analysis Techniques
| Technique |
Instrument Model |
| Liquid Chromatography-Mass Spectrometry (LC-MS) |
Thermo Scientific Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Q Exactive Plus) |
| Gas Chromatography-Mass Spectrometry (GC-MS) |
Agilent 7890A Gas Chromatograph coupled with Agilent 5975C Mass Selective Detector (Agilent 7890A/5975C GC-MS) |
| Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) |
Waters Xevo TQ-S Triple Quadrupole Mass Spectrometer (Waters Xevo TQ-S) |
| High-Resolution Mass Spectrometry (HRMS) |
Thermo Scientific Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Orbitrap Fusion Lumos) |
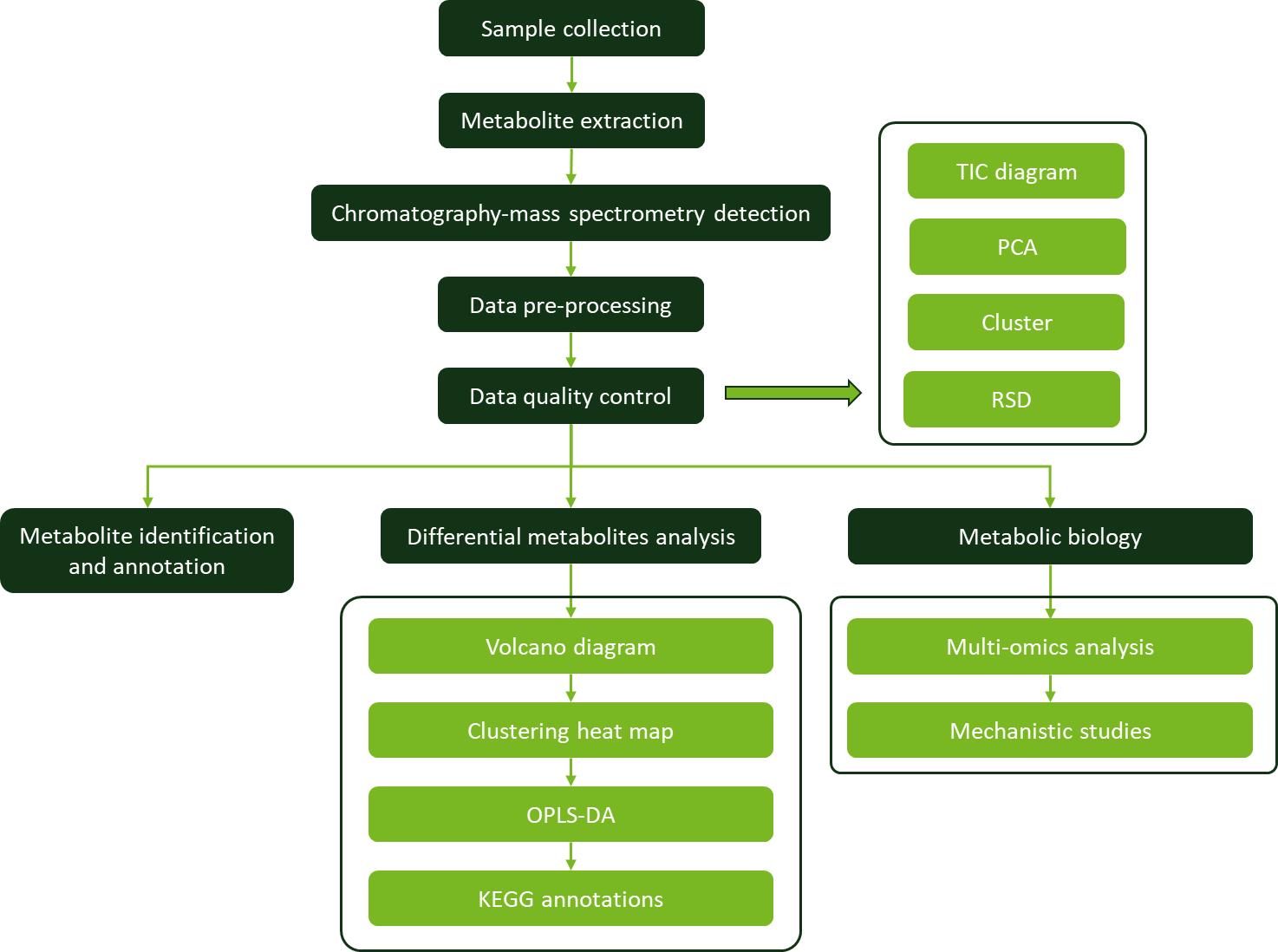 Workflow for Metabolomics Service
Workflow for Metabolomics Service
Sample Requirements for Common Sunflower Metabolomics
| Sample Type |
Sample Amount |
Sample Preparation |
Storage Conditions |
| Leaves |
50-100 mg |
Freeze-drying, grinding |
Store at -80°C |
| Roots |
50-100 mg |
Freeze-drying, grinding |
Store at -80°C |
| Seeds |
50-100 mg |
Grinding |
Store at -20°C in an airtight container |
| Flowers |
50-100 mg |
Freeze-drying, grinding |
Store at -80°C |
| Stem |
50-100 mg |
Freeze-drying, grinding |
Store at -80°C |
| Whole Plant (Tissue mix) |
50-100 mg |
Freeze-drying, grinding |
Store at -80°C |
| Extracts |
As per experiment |
Liquid-liquid extraction |
Store at -80°C |
| Cell Cultures |
As per experiment |
Harvest and extract |
Store at -80°C |
Case. Metabolomic Profiling of Sunflower Leaves Reveals Biomarkers for Drought Stress and Line Status
Background
This study aims to explore the metabolite composition of sunflower leaves and identify potential metabolic markers for distinguishing between different plant conditions and line statuses. The research focuses on the impact of drought stress on leaf metabolism and the ability to discriminate between restorer (R) and maintainer (B) sunflower lines.
Samples
The study utilizes adult sunflower leaves subjected to different conditions, including well-watered (WW) and drought-stressed (DS) samples. Additionally, it investigates metabolic differences between restorer (R) and maintainer (B) sunflower lines.
Technological Methods
Metabolite Quantification: The study quantifies 38 primary metabolites in sunflower leaves, including organic acids, fatty acids, amino acids, and soluble sugars, using various analytical techniques such as spectrophotometry, GC-FID, and UPLC-fluo.
1H-NMR Profiling: Polar extracts are analyzed using 1H-NMR to identify and quantify 20 annotated compounds in the millimolar range, providing additional insights into primary metabolism.
LC–ESI–QTOF–MS Analysis: Semi-polar extracts are analyzed using LC–ESI–QTOF–MS to detect specialized metabolites. Compounds with high intensities are tentatively annotated, including caffeoylquinic acids, methyl-flavonoids, and sesquiterpenoids.
Variable Selection Process: To reduce the number of variables for discrimination, three variable selection methods (ANOVA, sPLS, and LASSO penalty) are applied. This step helps identify the most relevant metabolic markers.
Results
Metabolite Composition: Sunflower leaves are characterized by their composition of major organic acids, fatty acids, amino acids, and soluble sugars. Specific compounds, such as malate, citrate, chlorogenic acid, and linolenic acid, are identified as prevalent in sunflower leaves.
Specialized Metabolites: LC–ESI–QTOF–MS analysis uncovers specialized metabolites, including caffeoylquinic acids, methyl-flavonoids, and sesquiterpenoids, which play various roles in sunflower physiology.
Biomarkers of Line Status: Metabolic markers help discriminate between restorer (R) and maintainer (B) sunflower lines. These markers include organic acids and unidentified ions measured by LC–ESI–QTOF–MS.
Biomarkers of Water Treatment: Metabolic markers efficiently distinguish between well-watered (WW) and drought-stressed (DS) sunflower samples. Amino acids, glycine-betaine, myo-inositol, caffeoylquinates, and sesquiterpenoids are identified as potential DS markers.
Small Efficient Biomarker Dataset: A set of eight biochemical variables, including total free amino acids, citrate, glycine-betaine, myo-inositol, sucrose, glucose, total proteins, and starch, is proposed as a cost-efficient metabolic marker set for discriminating between WW and DS samples. This set outperforms traditional physiological markers for assessing drought stress.
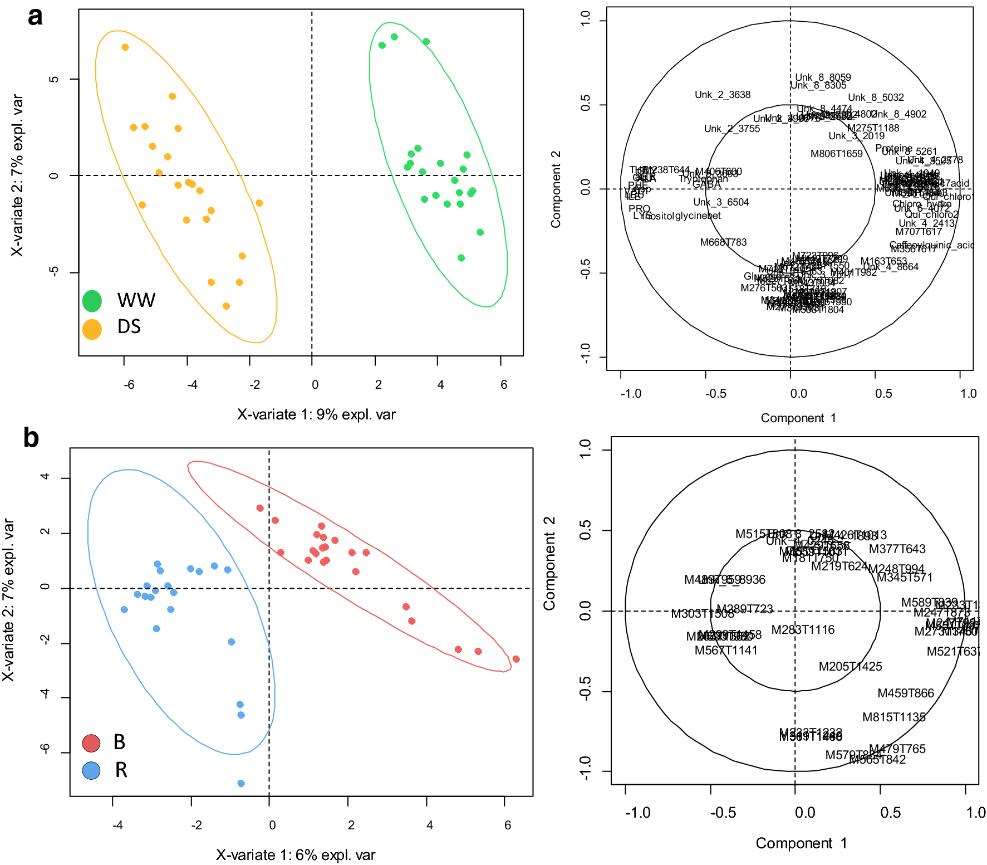 Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers
Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers
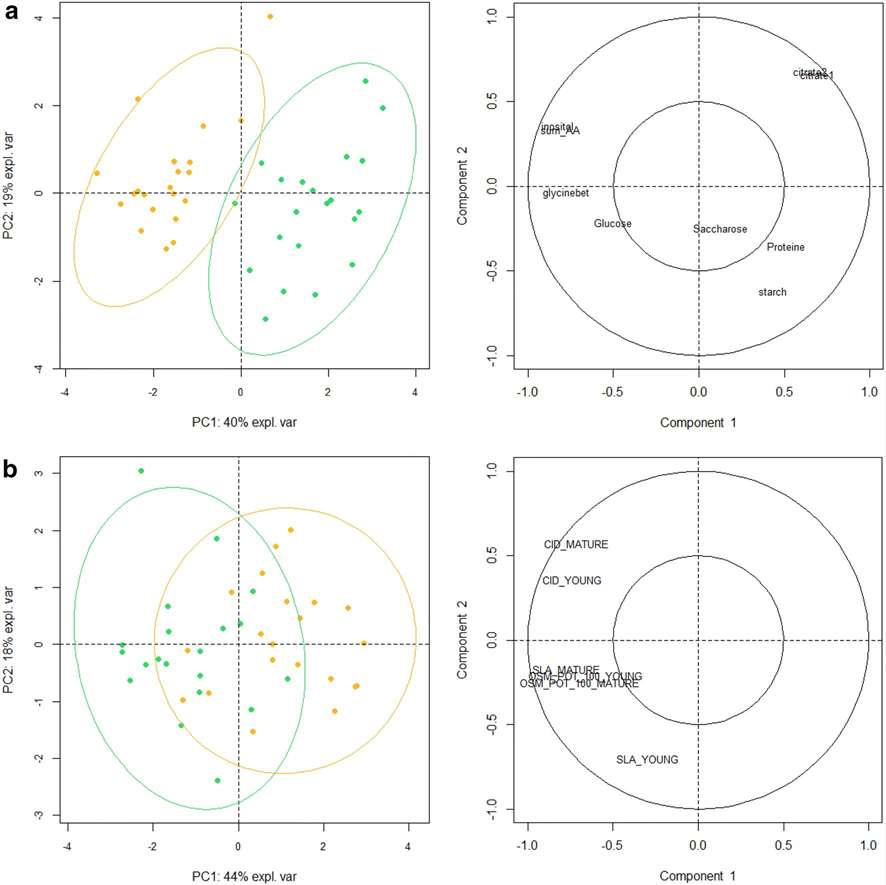 Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers
Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers
Reference
- Fernandez, Olivier, et al. "Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers." Metabolomics 15 (2019): 1-14.


 Workflow for Metabolomics Service
Workflow for Metabolomics Service Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers
Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers
Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers