What are Amino Acid Metabolites?
Amino acid metabolites are small organic molecules that are derived from the breakdown, synthesis, and interconversion of amino acids in various metabolic pathways. Amino acids are the building blocks of proteins and play crucial roles in numerous biological processes. Metabolites are the intermediates or end products of metabolic reactions.
Amino acid metabolites are involved in a wide range of biological functions. They participate in energy production, serving as substrates for cellular respiration and contributing to the synthesis of adenosine triphosphate (ATP), the universal energy currency of cells. Amino acid metabolites also serve as precursors for the synthesis of other important molecules, such as neurotransmitters, hormones, nucleotides, and lipids.
The metabolism of amino acids occurs through several interconnected pathways, including the catabolic pathways that involve the breakdown of amino acids for energy production and the anabolic pathways that involve the synthesis of new molecules from amino acids. During catabolism, amino acids are converted into various metabolites through processes such as deamination, decarboxylation, transamination, and oxidative reactions.
The specific amino acid metabolites generated depend on the metabolic state of the organism and the availability of enzymes and cofactors involved in these pathways. Examples of amino acid metabolites include alanine, glutamate, aspartate, γ-aminobutyric acid (GABA), ornithine, citrulline, and many others. Each metabolite has distinct biological functions and may serve as a signaling molecule, a precursor for the synthesis of other molecules, or a regulator of metabolic processes.
Analyzing amino acid metabolites provides valuable insights into the metabolic state and physiological conditions of organisms. Changes in amino acid metabolite profiles can serve as biomarkers for disease diagnosis, prognosis, and monitoring therapeutic interventions. Furthermore, studying amino acid metabolites helps in understanding the underlying mechanisms of metabolic disorders and facilitates the development of targeted treatments.
Amino Acid Metabolomics Service Provided by Creative Proteomics
Creative Proteomics utilizes a combination of GC/MS (Agilent 7890A/5975C) and LC/MS/MS (AB QTRAP 6500) platforms. This enables the quantitative and qualitative analysis of over 70 amino acids and their derivatives, 95% of which are quantified using standards and isotope standards.
Amino Acid Metabolome Profiling: Creative Proteomics offers comprehensive amino acid metabolome profiling services. Using advanced mass spectrometry-based techniques, they can analyze and quantify a wide range of amino acid metabolites in biological samples. This profiling allows for a holistic understanding of the metabolic state and dynamics of amino acids within the sample.
Targeted Amino Acid Analysis: For focused and precise analysis of specific amino acids or a subset of amino acids, Creative Proteomics provides targeted metabolomics services. These services utilize multiple reaction monitoring (MRM) techniques, which offer high sensitivity and specificity for the detection and quantification of selected amino acids in complex biological matrices.
Isotope Labeling Studies: Creative Proteomics specializes in stable isotope labeling studies for amino acid metabolism research. By incorporating stable isotopes into amino acids, they can trace the metabolic fate of amino acids and study the dynamics of metabolic pathways. Isotope labeling studies provide valuable insights into the utilization, interconversion, and turnover rates of amino acids.
Data Analysis and Interpretation: Creative Proteomics not only performs the experimental analysis but also offers comprehensive data analysis and interpretation services. Their team of experts can analyze the metabolomics data generated from amino acid metabolomics studies, apply statistical analysis, and provide meaningful insights and interpretations of the results.
Customized Solutions: Creative Proteomics understands that each research project has unique requirements. Therefore, they offer customized solutions tailored to specific research objectives. They collaborate closely with clients to design experiments, select appropriate analytical methods, optimize sample preparation protocols, and provide customized data analysis strategies.
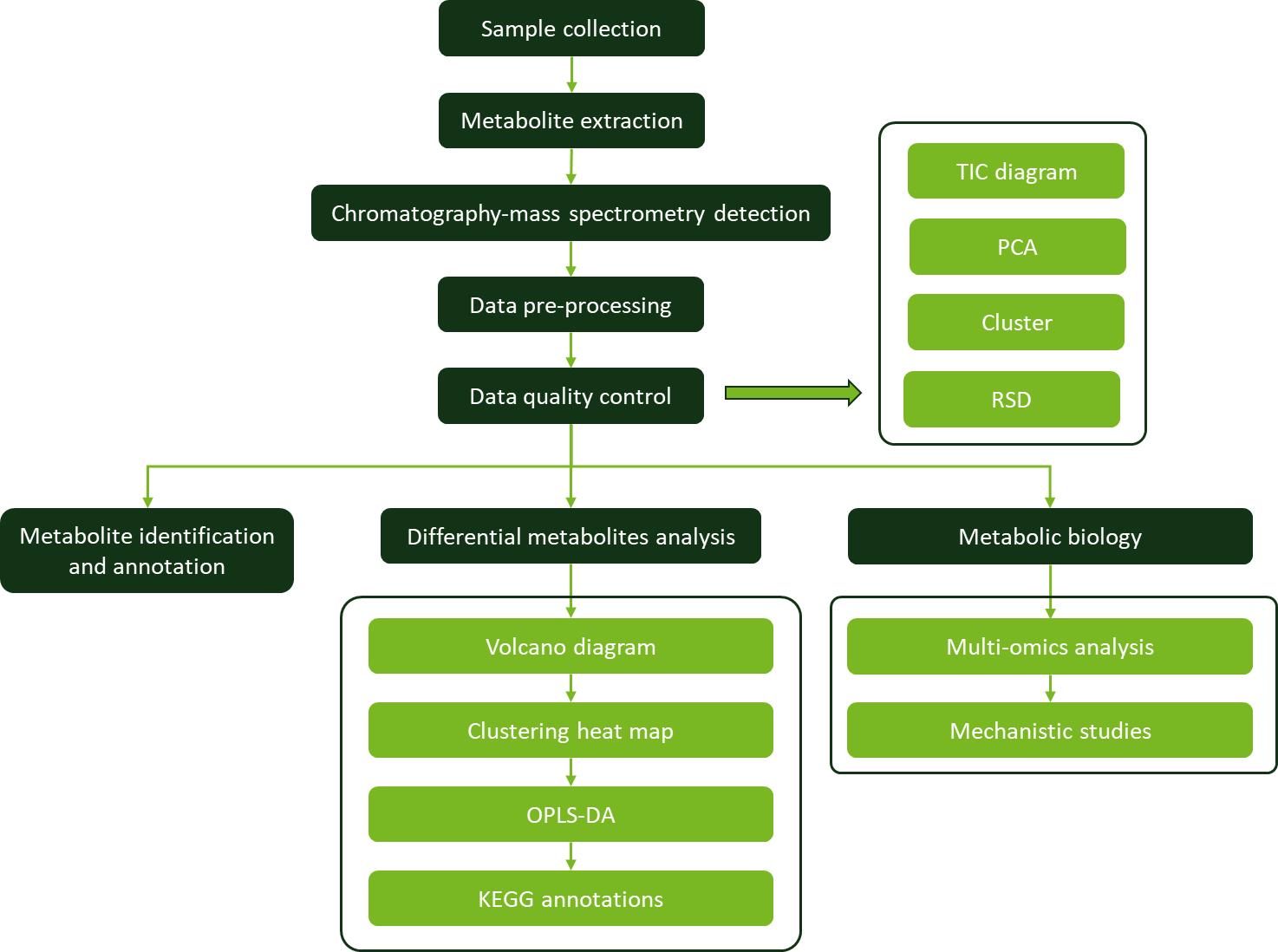 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
List of Amino Acid Metabolites and Derivatives Analyzed (including but not limited to)
| Alanine |
Arginine |
Asparagine |
Aspartic acid |
Cysteine |
| Glutamic acid |
Glutamine |
Glycine |
Histidine |
Isoleucine |
| Leucine |
Lysine |
Methionine |
Phenylalanine |
Proline |
| Serine |
Threonine |
Tyrosine |
Valine |
Tryptophan |
| Tyrosine |
Citrulline |
Ornithine |
Carnosine |
GABA (Gamma-aminobutyric acid) |
| Taurine |
Homocysteine |
Beta-alanine |
Agmatine |
Hydroxyproline |
| Creatine |
Hydroxyglutamate |
1-Methylhistidine |
3-Methylhistidine |
Phosphoserine |
| Delta-hydroxylysine |
Homocystine |
Alpha-aminoisobutyric acid |
Beta-aminoisobutyric acid |
|
Why Choose Us?
- Precise quantification: absolute quantification of internal standard, linear relationship of standard curve reaches more than 0.99.
- Wide database: covering 94 amino acids and their related substances.
- High sensitivity: using AB QTRAP® 6500+ LC-MS/MS, the sensitivity can reach ng/mL level.
Applications of Amino Acid Metabolomics
Plant Physiology: By analyzing changes in amino acid profiles, scientists can identify metabolic pathways involved in growth, development, and responses to environmental cues. This knowledge contributes to a deeper understanding of plant physiology and can be used to optimize agricultural practices.
Stress Response: Plants respond to various stressors such as drought, salinity, pathogens, and temperature fluctuations by altering their amino acid metabolism. By examining changes in amino acid levels and composition, researchers can identify key stress-responsive pathways and molecules. This information aids in developing stress-tolerant crop varieties and improving agricultural resilience.
Plant-Microbe Interactions: By studying the changes in amino acid profiles during these interactions, researchers can identify signaling molecules, defense mechanisms, and nutrient exchanges between plants and microbes.
Secondary Metabolite Production: Many secondary metabolites, including phytochemicals with medicinal properties, are derived from amino acid precursors. By analyzing amino acid metabolism, researchers can identify key enzymatic steps and regulatory mechanisms involved in secondary metabolite production. This information aids in improving plant-based pharmaceutical and nutraceutical production.
Crop Improvement: Amino acid metabolomics has the potential to enhance crop breeding and improvement programs. By studying amino acid profiles, researchers can identify markers associated with desirable traits such as yield, nutritional quality, and stress tolerance. This information facilitates marker-assisted selection and the development of improved crop varieties with enhanced agronomic traits.
Disease Research: By analyzing changes in amino acid levels and metabolism, scientists can identify metabolic dysregulations and gain insights into disease mechanisms.
Biomarker Discovery: By comparing the levels of amino acids and their metabolites in healthy individuals and patients, researchers can identify specific biomarkers that aid in disease diagnosis, prognosis, and monitoring.
Sample Requirements for Amino Acid Metabolites Assay
| Sample Types |
Minimum Sample Size |
Biological Repeat |
| Plant Tissues |
Stems, buds, nodes, leaves, flowers, fruits, roots, healing tissue |
600mg |
3-6 |
| Animal solids |
Whole insect body, flight apparatus (wings), nymphs, eggs, large pieces of animal tissue, red algae, cartilage, sclerotium |
500mg |
Humans >30/group
Animals 8-10/group |
| Animal liquids |
Plasma, serum, hemolymph, milk, egg white |
100 μL |
Humans >30/group
Animals 8-10/group |
| Cell samples |
Adherent cells |
1×107 |
Humans >30/group
Animals 8-10/group |
Case 1. Amino Acid Metabolism during Stress and Recovery in Arabidopsis: Insights into Physiological Responses and Toxicity
Background:
In the study, the researchers aimed to understand the behavior of amino acid metabolism during the recovery phase from high salinity and low water potential stress in Arabidopsis. Amino acids play crucial roles in plant stress responses, and their metabolism is intricately linked to plant growth, development, and stress tolerance. By analyzing the changes in amino acid metabolism, the researchers aimed to gain insights into the physiological and biochemical processes involved in stress and stress recovery.
Sample:
The samples used in the study were total leaf protein fractions isolated from Arabidopsis plants. These samples were collected at different time points, including before and after the stress treatment and at 6 and 18 hours of recovery. The samples were subjected to label-free shotgun proteomics to analyze the global response of the leaf proteome during stress and stress release.
Technical Methodology:
The determination of metabolite levels in the study involved specific procedures for amino acid quantification and proline quantification.
Amino Acid Quantification:
- Sample Preparation: Leaf material was lyophilized, and approximately 10 mg of plant powder was used for amino acid extraction.
- Extraction: The samples were solubilized in 0.1 M HCl and incubated at room temperature. After centrifugation, the supernatants were collected and stored.
- Derivatization: Before analysis, the extracts were diluted and mixed with internal standards. Precolumn derivatization was carried out using borate buffer, ortho-phthaldialdehyde, and 9-fluorenylmethoxycarbonyl chloride.
- UHPLC Analysis: The derivatized samples were analyzed using Ultra High-Performance Liquid Chromatography (UHPLC) with a specific gradient program and a C18 column. Amino acids were detected using a fluorescence detector and UV detector.
- Data Analysis: Peak areas of amino acids were evaluated using software.
Proline Quantification:
- Sample Preparation: Lyophilized plant powder (10 mg) was used for proline extraction.
- Extraction: The sample extract was prepared using sulfosalicylic acid and acidic ninhydrin.
- Reaction: The extract was incubated at 96°C in the dark, followed by stopping the reaction on ice.
- Separation: Toluene was added to the reaction mixture to separate the organic and water phases.
- Chromophore Measurement: The chromophore phase containing toluene was transferred to a quartz 96-well plate and read at 520 nm using a microplate reader.
- Calibration: The system was calibrated using known proline concentrations.
Results
Amino Acid Synthesis and Degradation:
The synthesis pathways for arginine and aromatic amino acids were significantly up-regulated during the recovery phase following both PEG and salt stress.
The abundance of enzymes involved in branched-chain amino acid biosynthesis was significantly decreased after stress.
The synthesis pathway for proline (Pro) was induced during both salt and PEG stress, with a stronger and more persistent response observed with PEG treatment.
The enzyme delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDH), involved in Pro degradation, was already induced during stress and remained at high levels during stress release.
Changes in Amino Acid Profiles:
The effect of salt stress on the leaf amino acid profile was less pronounced compared to PEG treatment. Only four amino acids significantly changed their contents after salt stress, while 10 were increased throughout the entire experiment during PEG stress.
The total free amino acid content increased during stress and showed a second peak during late recovery.
Glycine (Gly) was the only amino acid consistently lower in stressed plants compared to controls.
Glutamine (Gln) and alanine (Ala) strongly accumulated during the recovery period from PEG treatment.
Arginine (Arg) decreased significantly during recovery from PEG, possibly due to a high demand for polyamine synthesis, which was induced during stress.
Amino Acid Degradation and Toxicity:
The concentrations of low-abundant amino acids, including branched-chain amino acids, lysine (Lys), methionine (Met), histidine (His), and aromatic amino acids, showed the strongest relative increase after PEG treatment.
The synthesis pathways for these amino acids were generally down-regulated during stress and gradually returned to control levels during recovery.
The degradation pathways for these amino acids were strongly induced, peaking during early recovery before decreasing again.
A root growth inhibition assay revealed that Lys was the most toxic amino acid, inhibiting Arabidopsis root growth by 80% at a concentration of 100 μM, followed by leucine (Leu) and Met. Valine (Val) and isoleucine (Ile) showed mild toxic effects, while His addition slightly promoted root growth.
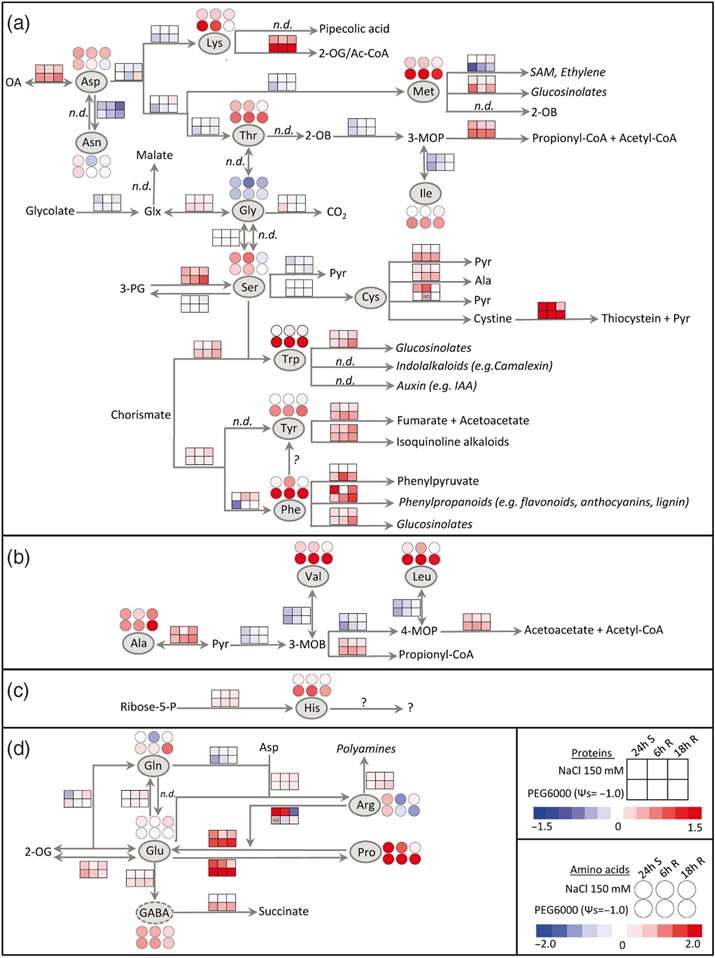 Amino acid metabolism during stress and stress release.
Amino acid metabolism during stress and stress release.
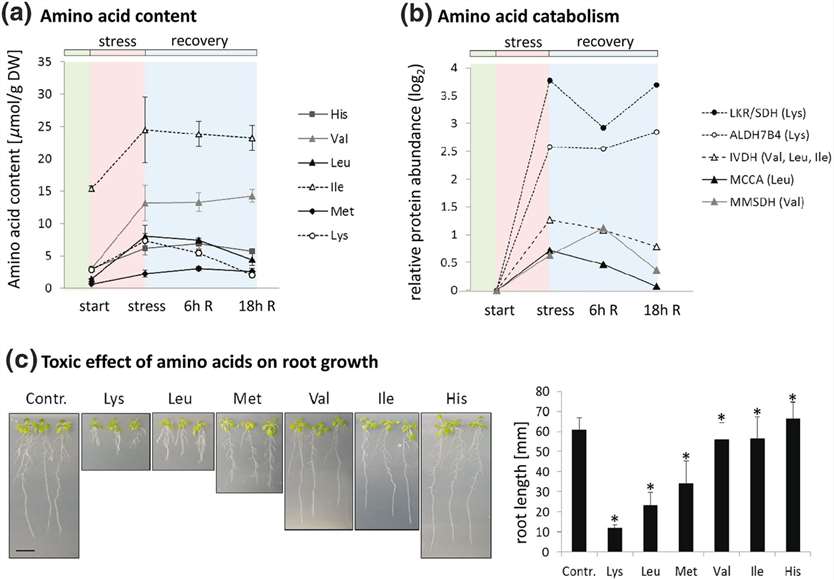 Rapid degradation of toxic amino acids during PEG stress and stress release
Rapid degradation of toxic amino acids during PEG stress and stress release
Reference
- Batista‐Silva, Willian, et al. "The role of amino acid metabolism during abiotic stress release." Plant, cell & environment 42.5 (2019): 1630-1644.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Amino acid metabolism during stress and stress release.
Amino acid metabolism during stress and stress release. Rapid degradation of toxic amino acids during PEG stress and stress release
Rapid degradation of toxic amino acids during PEG stress and stress release