What is Brassica oleracea?
Brassica oleracea is a highly diverse species encompassing a wide range of vegetables that have been cultivated for centuries. These vegetables are not only a staple in many diets but also renowned for their nutritional value and potential health benefits. Brassica oleracea cultivars are rich in phytochemicals, including glucosinolates, flavonoids, and phenolic compounds, which have been associated with various health-promoting properties such as antioxidant and anticancer activities.
Metabolomics, one of the omics sciences, focuses on the comprehensive analysis of small molecules (metabolites) in biological systems. When applied to Brassica oleracea, metabolomics offers several compelling advantages:
- Holistic Understanding: Metabolomics provides a holistic view of the plant's metabolic profile, allowing researchers to understand the intricate biochemical processes underlying its growth, development, and response to environmental factors.
- Nutritional Insights: Brassica oleracea cultivars are valued for their nutritional content. Metabolomics helps uncover the specific metabolites responsible for their health benefits and allows for targeted breeding to enhance these traits.
- Quality Control: Metabolomics aids in quality control of Brassica oleracea-based products, ensuring consistency and purity in food and pharmaceutical industries.
Creative Proteomics offers Brassica oleracea metabolomics analysis services, providing comprehensive solutions for the in-depth study of this versatile plant species. With years of experience, a team of top biologists, and advanced instrumentation, we specialize in sample preparation, state-of-the-art analytical techniques, and advanced data analysis to uncover the metabolic secrets of Brassica oleracea, whether for research into growth and development or exploring its potential health and economic value.
Our Brassica oleracea Metabolomics Analysis Projects
1. Cotton Plant-Microbe Interactions
Understanding the intricate relationships between cotton plants and various microbes is crucial for improving cotton crop health and productivity. Our research in this area delves into the metabolic responses of cotton to microbial interactions, providing insights that help develop strategies for enhanced disease resistance and yield optimization.
2. Stress Responses in Cotton
Cotton plants face environmental stressors, including drought, extreme temperatures, and salinity. Our metabolomics analysis aids in uncovering the metabolic pathways and key metabolites involved in stress tolerance. By identifying and targeting these specific metabolites, we assist in developing cotton varieties with superior stress resilience.
3. Phytochemical Mediators
We specialize in the identification and quantification of phytochemicals in cotton plants. This information supports the development of sustainable pest control strategies based on naturally occurring compounds, reducing the reliance on chemical pesticides and promoting environmentally friendly approaches in cotton cultivation.
4. Nutritional Enhancement
Our research is dedicated to enhancing the nutritional value of cottonseeds. By identifying and quantifying specific metabolites, such as essential fatty acids and proteins, we aim to improve the nutritional content of cottonseeds, ultimately increasing their value for various applications, including animal feed and food products.
5. Development of Cotton Varieties
Creative Proteomics has actively contributed to the development of specialized cotton varieties. Through metabolomics analysis, we can select and breed cultivars with specific metabolic profiles, tailored to meet various industrial or nutritional requirements. This approach contributes to the creation of cotton varieties that are high-yielding and customized for specific applications.
6. Fiber Development and Quality
We focus on the metabolic processes governing cotton fiber development and quality. By identifying metabolites associated with superior fiber characteristics, such as length, strength, and fineness, we contribute to the production of cotton with the highest-quality fibers, meeting the demands of the textile market.
7. Sustainable Agriculture Practices
Sustainability is a paramount concern in modern agriculture. Our research explores the metabolic changes in cotton plants under sustainable agricultural practices, such as organic farming or reduced pesticide usage. This research helps identify metabolic markers associated with sustainable cotton cultivation.
8. Environmental Stressors and Cotton Adaptation
In a changing climate, cotton plants are exposed to various environmental stressors. Our metabolomics analysis is instrumental in understanding how cotton plants adapt to these changing conditions, informing strategies for cultivating cotton varieties that can thrive in diverse climatic conditions.
9. Cottonseed Oil Composition
Cottonseed oil is a valuable commodity with applications in the food and industrial sectors. We examine the composition of cottonseed oil, with a focus on fatty acid profiles. By identifying and quantifying specific fatty acids, we contribute to the development of cotton varieties with tailored oil compositions for various applications.
10. Disease Resistance and Biocontrol
Cotton is susceptible to various diseases, and sustainable disease management is essential for crop protection. Our research explores the metabolic responses of cotton plants to diseases and the interactions with beneficial microorganisms, supporting the development of integrated pest and disease management strategies for cotton.
Analytical Techniques for Brassica oleracea Metabolomics
Liquid chromatography-mass spectrometry (LC-MS) is a cornerstone of our analysis. High-resolution LC-MS instruments, notably the Thermo Fisher Q-Exactive series, are employed. These cutting-edge instruments are chosen for their remarkable sensitivity and resolution, enabling the detection of a diverse range of metabolites.
Gas Chromatography-Mass Spectrometry (GC-MS) is our second pivotal technique. In this method, instruments like the Agilent 7890A GC coupled to the Agilent 5975C MS are employed. GC-MS is instrumental in the analysis of volatile and thermally stable metabolites.
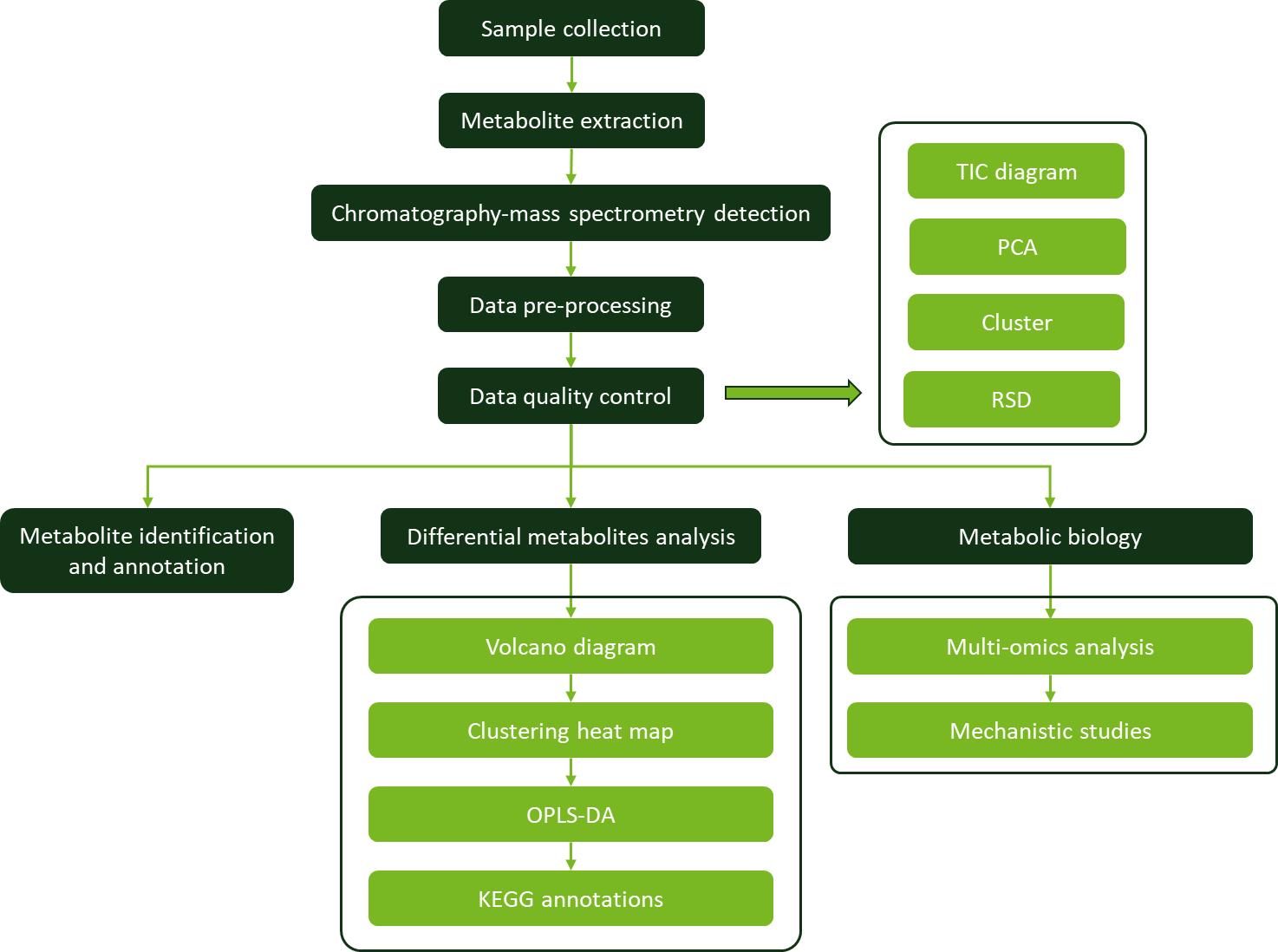 Workflow for Metabolomics Service
Workflow for Metabolomics Service
Sample Requirements for Brassica oleracea Metabolomics
| Sample Type |
Suggested Sample Quantity |
| Roots |
50-100 grams (fresh weight) |
| Stems and Leaves |
50-100 grams (fresh weight) |
| Flowers |
5-10 grams (fresh weight) |
| Fruits/Seeds |
20-50 grams (fresh weight) |
| Rhizomes |
20-50 grams (fresh weight) |
| Shoots and Young Leaves |
50-100 grams (fresh weight) |
| Tissue Sections |
Variable based on study needs |
| Seedlings/Saplings |
20-50 grams (fresh weight) |
| Rhizosphere Soil |
100-200 grams (dry weight) |
| Root Exudates |
Variable based on study needs |


 Workflow for Metabolomics Service
Workflow for Metabolomics Service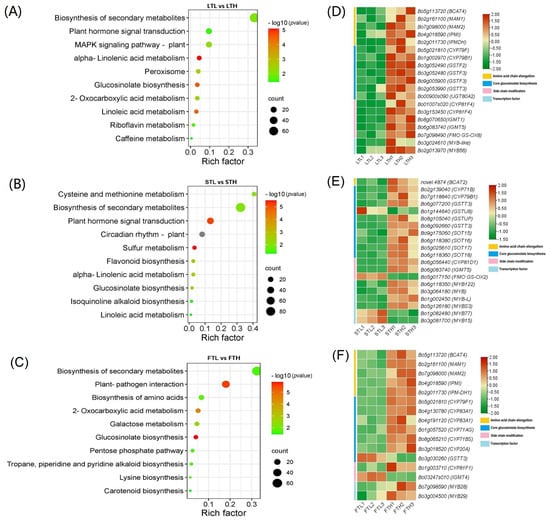 KEGG pathway enrichment analyses of DEGs identified between low and high RAA content pools.
KEGG pathway enrichment analyses of DEGs identified between low and high RAA content pools.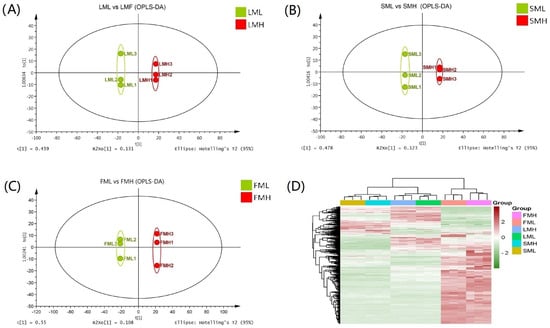 Metabolomics profiling between low and high RAA content pools.
Metabolomics profiling between low and high RAA content pools.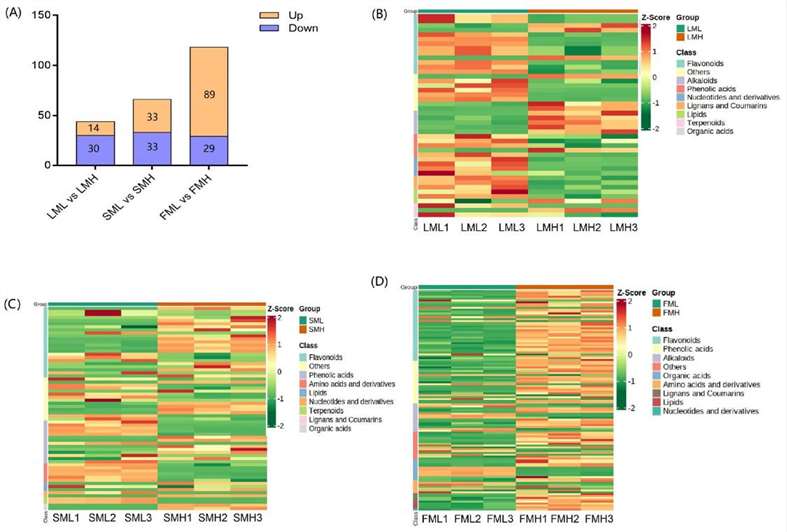 Differential metabolites analysis.
Differential metabolites analysis.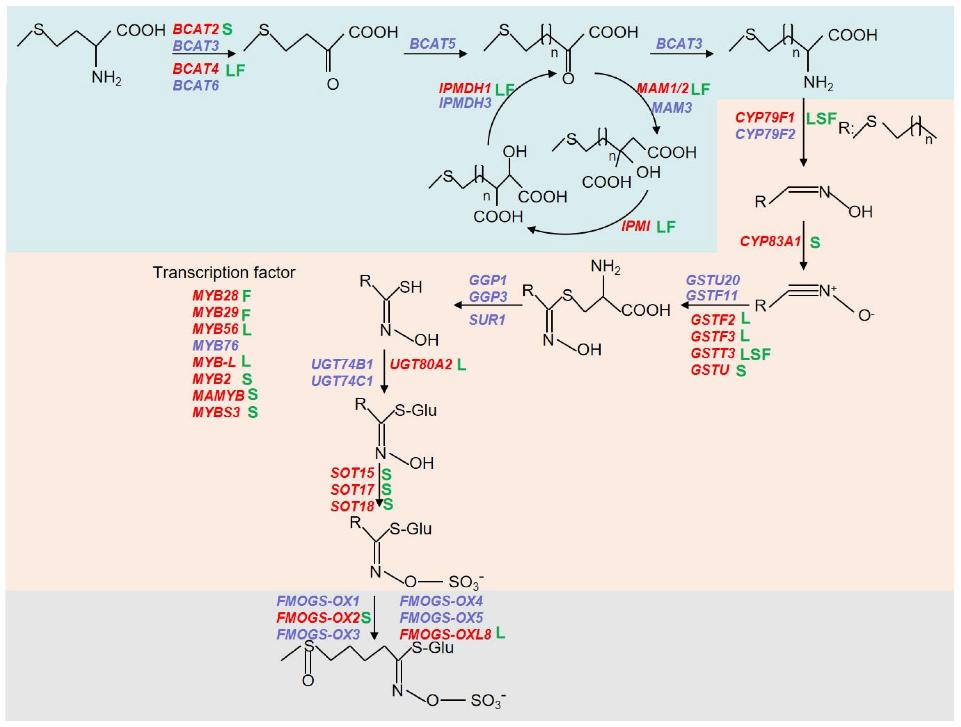 Schematic biosynthetic pathway of aliphatic glucosinolates and involved genes.
Schematic biosynthetic pathway of aliphatic glucosinolates and involved genes.